Opening Pandora's Box: High-Level Resistance to Antibiotics of Last
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Factors Affecting Effective Teaching and Learning of Economics in Some Ogbomosho High Schools, Oyo State, Nigeria
Journal of Education and Practice www.iiste.org ISSN 2222-1735 (Paper) ISSN 2222-288X (Online) Vol.7, No.28, 2016 Factors Affecting Effective Teaching and Learning of Economics in Some Ogbomosho High Schools, Oyo State, Nigeria Gbemisola Motunrayo Ojo Vusy Nkoyane Department of Curriculum and Instructional Studies, College of Education, University of South Africa 0001 Abstract This study was carried out to examine the present curriculum of Economics as a subject in some Ogbomoso Senior High Schools and to determine factors affecting effective teaching of economics in the schools. Variables such as number of students, teachers’ ratio available textbooks were also examined. The study adopted descriptive design since it is an ex post facto research. The target population for this study is Ogbomosho North Local Government Area of Oyo State comprising nine (9) High Secondary Schools. Three (3) teachers of economics and five (5) students from each school were used in answering the questionnaires. The study revealed high number of economics students (5,864) as against 26 teachers in nine (9) public schools under study. This gives a very high teacher-student ratio of 1:225. The findings also showed that there one lack of teaching aids, library facilities and where available, there is lack of textbooks of Economics. All the schools studied were highly dense resulted into high students’ population. Based on the findings of his study, the federal and the state governments should employ more teachers, especially in Economics to checkmate rising teacher student ratio. More senior secondary schools should be built with the increasing student population within the LGA under study. -
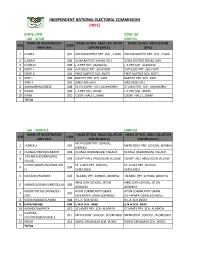
State: Oyo Code: 30 Lga : Afijio Code: 01 Name of Registration Name of Reg
INDEPENDENT NATIONAL ELECTORAL COMMISSION (INEC) STATE: OYO CODE: 30 LGA : AFIJIO CODE: 01 NAME OF REGISTRATION NAME OF REG. AREA COLLATION NAME OF REG. AREA CENTRE S/N CODE AREA (RA) CENTRE (RACC) (RAC) 1 ILORA I 001 OKEDIJI BAPTIST PRY. SCH., ILORA OKEDIJI BAPTIST PRY. SCH., ILORA 2 ILORA II 002 ILORA BAPTIST GRAM. SCH. ILORA BAPTIST GRAM. SCH. 3 ILORA III 003 L.A PRY SCH. ALAWUSA. L.A PRY SCH. ALAWUSA. 4 FIDITI I 004 CATHOLIC PRY. SCH FIDITI CATHOLIC PRY. SCH FIDITI 5 FIDITI II 005 FIRST BAPTIST SCH. FIDITI FIRST BAPTIST SCH. FIDITI 6 AWE I 006 BAPTIST PRY. SCH. AWE BAPTIST PRY. SCH. AWE 7 AWE II 007 AWE HIGH SCH. AWE HIGH SCH. 8 AKINMORIN/JOBELE 008 ST.JOHN PRY. SCH. AKINMORIN ST.JOHN PRY. SCH. AKINMORIN 9 IWARE 009 L.A PRY SCH. IWARE. L.A PRY SCH. IWARE. 10 IMINI 010 COURT HALL 1, IMINI COURT HALL 1, IMINI TOTAL LGA : AKINYELE CODE: 02 NAME OF REGISTRATION NAME OF REG. AREA COLLATION NAME OF REG. AREA COLLATION S/N CODE AREA (RA) CENTRE (RACC) CENTRE (RACC) METHODIST PRY. SCHOOL, 1 IKEREKU 001 METHODIST PRY. SCHOOL, IKEREKU IKEREKU 2 OLANLA/OBODA/LABODE 002 OLANLA (OGBANGAN) VILLAGE OLANLA (OGBANGAN) VILLAGE EOLANLA (OGBANGAN) 3 003 COURT HALL ARULOGUN VILLAGE COURT HALL ARULOGUN VILLAGE VILLAG OLODE/AMOSUN/ONIDUND ST. LUKES PRY. SCHOOL, ST. LUKES PRY. SCHOOL, 4 004 U ONIDUNDU ONIDUNDU 5 OJO-EMO/MONIYA 005 ISLAMIC PRY. SCHOOL, MONIYA ISLAMIC PRY. SCHOOL, MONIYA ANGLICAN SCHOOL, OTUN ANGLICAN SCHOOL, OTUN 6 AKINYELE/ISABIYI/IREPODUN 006 AGBAKIN AGBAKIN IWOKOTO/TALONTAN/IDI- AYUN COMMUNITY GRAM. -

Odo/Ota Local Government Secretariat, Sango - Agric
S/NO PLACEMENT DEPARTMENT ADO - ODO/OTA LOCAL GOVERNMENT SECRETARIAT, SANGO - AGRIC. & BIO. ENGINEERING 1 OTA, OGUN STATE AGEGE LOCAL GOVERNMENT, BALOGUN STREET, MATERNITY, AGRIC. & BIO. ENGINEERING 2 SANGO, AGEGE, LAGOS STATE AHMAD AL-IMAM NIG. LTD., NO 27, ZULU GAMBARI RD., ILORIN AGRIC. & BIO. ENGINEERING 3 4 AKTEM TECHNOLOGY, ILORIN, KWARA STATE AGRIC. & BIO. ENGINEERING 5 ALLAMIT NIG. LTD., IBADAN, OYO STATE AGRIC. & BIO. ENGINEERING 6 AMOULA VENTURES LTD., IKEJA, LAGOS STATE AGRIC. & BIO. ENGINEERING CALVERTON HELICOPTERS, 2, PRINCE KAYODE, AKINGBADE MECHANICAL ENGINEERING 7 CLOSE, VICTORIA ISLAND, LAGOS STATE CHI-FARM LTD., KM 20, IBADAN/LAGOS EXPRESSWAY, AJANLA, AGRIC. & BIO. ENGINEERING 8 IBADAN, OYO STATE CHINA CIVIL ENGINEERING CONSTRUCTION CORPORATION (CCECC), KM 3, ABEOKUTA/LAGOS EXPRESSWAY, OLOMO - ORE, AGRIC. & BIO. ENGINEERING 9 OGUN STATE COCOA RESEARCH INSTITUTE OF NIGERIA (CRIN), KM 14, IJEBU AGRIC. & BIO. ENGINEERING 10 ODE ROAD, IDI - AYANRE, IBADAN, OYO STATE COKER AGUDA LOCAL COUNCIL, 19/29, THOMAS ANIMASAUN AGRIC. & BIO. ENGINEERING 11 STREET, AGUDA, SURULERE, LAGOS STATE CYBERSPACE NETWORK LTD.,33 SAKA TIINUBU STREET. AGRIC. & BIO. ENGINEERING 12 VICTORIA ISLAND, LAGOS STATE DE KOOLAR NIGERIA LTD.,PLOT 14, HAKEEM BALOGUN STREET, AGRIC. & BIO. ENGINEERING OPP. TECHNICAL COLLEGE, AGIDINGBI, IKEJA, LAGOS STATE 13 DEPARTMENT OF PETROLEUM RESOURCES, 11, NUPE ROAD, OFF AGRIC. & BIO. ENGINEERING 14 AHMAN PATEGI ROAD, G.R.A, ILORIN, KWARA STATE DOLIGERIA BIOSYSTEMS NIGERIA LTD, 1, AFFAN COMPLEX, 1, AGRIC. & BIO. ENGINEERING 15 OLD JEBBA ROAD, ILORIN, KWARA STATE Page 1 SIWES PLACEMENT COMPANIES & ADDRESSES.xlsx S/NO PLACEMENT DEPARTMENT ESFOOS STEEL CONSTRUCTION COMPANY, OPP. SDP, OLD IFE AGRIC. & BIO. ENGINEERING 16 ROAD, AKINFENWA, EGBEDA, IBADAN, OYO STATE 17 FABIS FARMS NIGERIA LTD., ILORIN, KWARA STATE AGRIC. -

Socio-Economic Impacts of Settlers in Ado-Ekiti
International Journal of History and Cultural Studies (IJHCS) Volume 3, Issue 2, 2017, PP 19-26 ISSN 2454-7646 (Print) & ISSN 2454-7654 (Online) DOI: http://dx.doi.org/10.20431/2454-7654.0302002 www.arcjournals.org Socio-Economic Impacts of Settlers in Ado-Ekiti Adeyinka Theresa Ajayi (PhD)1, Oyewale, Peter Oluwaseun2 Department of History and International Studies, Ekiti State University, Ado-Ekiti, Ekiti State Nigeria Abstract: Migration and trade are two important factors that led to the commercial growth and development of Ado-Ekiti during pre-colonial, colonial and post colonial period. These twin processes were facilitated by efforts of other ethnic groups, most notably the Ebira among others, and other Yoruba groups. Sadly there is a paucity of detailed historical studies on the settlement pattern of settlers in Ado-Ekiti. It is in a bid to fill this gap that this paper analyses the settlement pattern of settlers in Ado-Ekiti. The paper also highlights the socio- economic and political activities of settlers, especially their contributions to the development of Ado-Ekiti. Data for this study were collected via oral interviews and written sources. The study highlights the contributions of settlers in Ado-Ekiti to the overall development of the host community 1. INTRODUCTION Ado-Ekiti, the capital of Ekiti state is one of the state in southwest Nigeria. the state was carved out from Ondo state in 1996, since then, the state has witness a tremendous changes in her economic advancement. The state has witness the infix of people from different places. Settlement patterns in intergroup relations are assuming an important area of study in Nigeria historiography. -

A Perception Study of Development Control Activities in Ibadan, Osogbo and Ado-Ekiti
A Service of Leibniz-Informationszentrum econstor Wirtschaft Leibniz Information Centre Make Your Publications Visible. zbw for Economics Olowoporoku, Oluwaseun; Daramola, Oluwole; Agbonta, Wilson; Ogunleye, John Article Urban jumble in three Nigerian cities: A perception study of development control activities in Ibadan, Osogbo and Ado-Ekiti Economic and Environmental Studies (E&ES) Provided in Cooperation with: Opole University Suggested Citation: Olowoporoku, Oluwaseun; Daramola, Oluwole; Agbonta, Wilson; Ogunleye, John (2017) : Urban jumble in three Nigerian cities: A perception study of development control activities in Ibadan, Osogbo and Ado-Ekiti, Economic and Environmental Studies (E&ES), ISSN 2081-8319, Opole University, Faculty of Economics, Opole, Vol. 17, Iss. 4, pp. 795-811, http://dx.doi.org/10.25167/ees.2017.44.10 This Version is available at: http://hdl.handle.net/10419/193043 Standard-Nutzungsbedingungen: Terms of use: Die Dokumente auf EconStor dürfen zu eigenen wissenschaftlichen Documents in EconStor may be saved and copied for your Zwecken und zum Privatgebrauch gespeichert und kopiert werden. personal and scholarly purposes. Sie dürfen die Dokumente nicht für öffentliche oder kommerzielle You are not to copy documents for public or commercial Zwecke vervielfältigen, öffentlich ausstellen, öffentlich zugänglich purposes, to exhibit the documents publicly, to make them machen, vertreiben oder anderweitig nutzen. publicly available on the internet, or to distribute or otherwise use the documents in public. Sofern die Verfasser die Dokumente unter Open-Content-Lizenzen (insbesondere CC-Lizenzen) zur Verfügung gestellt haben sollten, If the documents have been made available under an Open gelten abweichend von diesen Nutzungsbedingungen die in der dort Content Licence (especially Creative Commons Licences), you genannten Lizenz gewährten Nutzungsrechte. -

Gastroesophageal Reflux Disease and Etiological Correlates Among Nigerian Adults at Ogbomoso A.C
Ann Ibd. Pg. Med 2018. Vol.16, No.1 30-36 GASTROESOPHAGEAL REFLUX DISEASE AND ETIOLOGICAL CORRELATES AMONG NIGERIAN ADULTS AT OGBOMOSO A.C. Jemilohun1, B.O. Oyelade2, J.O. Fadare3 and I.O. Amole4 1. Department of Medicine, Ladoke Akintola University of Technology/Ladoke Akintola University of Technology Teaching Hospital, Ogbomoso , Oyo State, Nigeria 2. Ladoke Akintola University of Technology Health Centre, Ogbomoso, Oyo State, Nigeria 3. Department of Pharmacology, Ekiti State University, Ado-Ekiti, Ekiti State, Nigeria 4. Department of Family Medicine, Bowen University Teaching Hospital, Ogbomoso, Oyo State, Nigeria Correspondence: ABSTRACT Dr. A.C. Jemilohun Background: Gastroesophageal reflux disease is a common disorder globally but LAUTECH Teaching Hospital, the prevalence and severity of symptoms and complications do have ethnic and Ogbomoso, Oyo State, geographic variations. Nigeria Aim: The aim of the study was to determine the prevalence of gastroesophageal Email: [email protected] reflux disease (GERD) among adults in Ogbomoso, Southwest Nigeria, and the risk factors associated with it. Methodology: The study was a hospital-based descriptive cross-sectional survey. The study population consisted of consenting male and female adults aged 18 to 87 years recruited from the outpatient departments of three health institutions in Ogbomoso, Oyo State, Nigeria. A predesigned questionnaire consisting of GerdQ and other relevant information was administered to the participants. Results: A total of 772 subjects were interviewed. The prevalence of GERD was 9.3% among the subjects. There was a significant association between increasing age and GERD. Other risk factors considered showed no significant association with the disease. Conclusion: GERD has a moderate prevalence among adults in Ogbomoso, Southwest Nigeria and there is a significant association between increasing age and GERD. -

IJHG May-Aug 07.Indd
59 Original Article Human sex ratio at birth in South West Nigeria M. A. Azeez*, A. Akinboro*^, A. A. Bakare^ Department of *Pure and Applied Biology, Ladoke Akintola University of Technology, P. M. B. 4000, Ogbomoso; ^Cell Biology and Genetics Unit, Department of Zoology, University of Ibadan, Ibadan. Nigeria The sex ratio of man is expected to be more or less equal BACKGROUND: Human sex ratio at birth differs from one population to the other. This variation has been attributed because of the 1:1 segregation of X and Y-chromosomes to cultural practices, seasonal variation, small-family in the male.[2] In 2004, males accounted for 50.4% of the size policy and sex selective technology. Information on global total.[3] In the absence of manipulation, human secondary sex ratio in Nigeria is limited. AIMS AND OBJECTIVE: To analyzed human sex ratio secondary sex ratio is commonly assumed to be 105- at birth for samples of the Nigerian population in 4 urban 107 male births for every 100 female birth. However, settings in Southwest Nigeria, in order to know the trend and to compare the fi ndings with those of previous reports. sex ratios at birth or among infants may be considerably MATERIALS AND METHODS: Data were collected from skewed by sex-selective abortion and infanticide. Even Obafemi Awolowo University (OAU) teaching hospital at Ile in the absence of such practices, a range of sex ratios Ife and Wesley Guild hospital at Ilesa, Osun state; General hospital at Ogbomoso, Oyo state and Ekiti state specialist at birth of between 103 to 107 boys per 100 girls has hospital at Ado-Ekiti, Ekiti state. -

Interpretation of Groundmagnetic Data in Oke-Ogba Area, Akure, Southwestern Nigeria
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Covenant University Repository Available online a t www.pelagiaresearchlibrary.com Pelagia Research Library Advances in Applied Science Research, 2012, 3 (5):3216-3222 ISSN: 0976-8610 CODEN (USA): AASRFC Interpretation of groundmagnetic data in Oke-Ogba Area, Akure, Southwestern Nigeria 1Sunmonu Lukman Ayobami, 1* Adagunodo Theophilus Aanuoluwa, 1Olafisoye Emmanuel Rotimi and 2Oladejo Olagoke Peter 1Department of Pure and Applied Physics, Ladoke Akintola University of Technology, P.M.B. 4000, Ogbomoso, Oyo State, Nigeria. 2Department of Physics, Emmanuel Alayande College of Education, Oyo, Oyo State, Nigeria. _____________________________________________________________________________________________ ABSTRACT Groundmagnetic method was used to delineate into the subsurface in Oke-Ogba area, Akure, which falls within latitude 07 0 14 ′ to 07 0 19 ′ North and longitude 005 0 06 ′ to 005 0 12 ′ East Southwestern Nigeria, with a view to determining the competent areas for engineering purposes and the promising areas for hydrogeologic prospects. A total of four traverses were established for the purpose of the groundmagnetic study, and the results were presented as groundmagnetic profiles of varying magnetic intensities. The result of investigation revealed that the intensities and characteristics nature of the magnetic anomalies as expressed by all profiles are indicative of the different rock types producing them. Qualitative and quantitative interpretation of individual magnetic anomaly and geological knowledge of the survey area yielded information on the depth of geological features (e.g. rock contact, faults or fractures), structures and magnetic properties of rock units. From the preliminary interpretation, the existence of some structural features such as a likely fault, fracture and contacts between rocks as well as relatively mineralized zones that might contain magnetic minerals of essential magnetite origin is revealed. -

Available Stanbic IBTC Bank Branches
Available Stanbic IBTC Bank Branches BRANCH OPENING OPENING S/N State Branch Name Branch Location CITY STATUS TIME 7, ABA-OWERRI ROAD P.M.B 7477, DAILY: 9AM - 1 ABIA ABA MAIN ABA ABA OPENED 3PM 7 DURU STREET OFF CEMETARY DAILY: 9AM - 2 ABIA ABA MARKET ROAD ABA OPENED 3PM 189, FAULKS ROAD, ARIARIA DAILY: 9AM - 3 ABIA ARIARIA MINI MARKET, ABA ABA OPENED 3PM 2 MARKET ROAD BY LIBRARY DAILY: 9AM - 4 ABIA UMUAHIA AVENUE, UMUAHIA UMUAHIA OPENED 3PM ABUJA SERVICE NO. 75, RALPH SHODEINDE DAILY: 9AM - 5 ABUJA CENTRE STREET ABUJA OPENED 2PM NNPC TOWER, HERBERT DAILY: 9AM - 6 ABUJA ABUJA-NNPC MACAULAY WAY, GARKI, ABUJA ABUJA OPENED 2PM AHMADU BELLO PLOT 149 AHMADU BELLO WAY, DAILY: 9AM - 7 ABUJA WAY AREA 11 GARKI ABUJA ABUJA OPENED 2PM DEIDE MARKET ROAD, OPPOSITE DAILY: 9AM - 8 ABUJA DEIDE MINI THE MARKET, DEIDE, ABUJA ABUJA OPENED 2PM INFINITY HOUSE, 11 KAURA DAILY: 9AM - 9 ABUJA GARKI (AREA 3) NAMODA STREET ABUJA OPENED 2PM GARKI MODEL PLOT CBN 2, LADOKE AKINTOLA DAILY: 9AM - 10 ABUJA MINI BLVD, ABUJA OPENED 2PM PLOT 415, SPECIALIST HOSPITAL DAILY: 9AM - 11 ABUJA GWAGWALADA ROAD GWAGWALADA GWAGWALADA OPENED 3PM 84/85 GADO NASKO WAY, KUBWA, DAILY: 9AM - 12 ABUJA KUBWA MINI ABUJA Kubwa OPENED 3PM PLOT 2777, CADASTRAL ZONE A6 DAILY: 9AM - 13 ABUJA MAITAMA P.M.B 337, ABUJA ABUJA OPENED 2PM NIGERIAN IMMIGRATION NIGERIAN IMMIGRATION DAILY: 9AM - 14 ABUJA SERVICE HEADQUARTERS, AIRPORT ROAD ABUJA OPENED 2PM SHOP 10, GRAND TOWER. ABUJA 15 ABUJA TOWER MALL MALL, APO, ABUJA ABUJA CLOSED CLOSED PLOT 37, EKUKINAM STREET, DAILY: 9AM - 16 ABUJA UTAKO BRANCH UTAKO ABUJA. -

States and Lcdas Codes.Cdr
PFA CODES 28 UKANEFUN KPK AK 6 CHIBOK CBK BO 8 ETSAKO-EAST AGD ED 20 ONUIMO KWE IM 32 RIMIN-GADO RMG KN KWARA 9 IJEBU-NORTH JGB OG 30 OYO-EAST YYY OY YOBE 1 Stanbic IBTC Pension Managers Limited 0021 29 URU OFFONG ORUKO UFG AK 7 DAMBOA DAM BO 9 ETSAKO-WEST AUC ED 21 ORLU RLU IM 33 ROGO RGG KN S/N LGA NAME LGA STATE 10 IJEBU-NORTH-EAST JNE OG 31 SAKI-EAST GMD OY S/N LGA NAME LGA STATE 2 Premium Pension Limited 0022 30 URUAN DUU AK 8 DIKWA DKW BO 10 IGUEBEN GUE ED 22 ORSU AWT IM 34 SHANONO SNN KN CODE CODE 11 IJEBU-ODE JBD OG 32 SAKI-WEST SHK OY CODE CODE 3 Leadway Pensure PFA Limited 0023 31 UYO UYY AK 9 GUBIO GUB BO 11 IKPOBA-OKHA DGE ED 23 ORU-EAST MMA IM 35 SUMAILA SML KN 1 ASA AFN KW 12 IKENNE KNN OG 33 SURULERE RSD OY 1 BADE GSH YB 4 Sigma Pensions Limited 0024 10 GUZAMALA GZM BO 12 OREDO BEN ED 24 ORU-WEST NGB IM 36 TAKAI TAK KN 2 BARUTEN KSB KW 13 IMEKO-AFON MEK OG 2 BOSARI DPH YB 5 Pensions Alliance Limited 0025 ANAMBRA 11 GWOZA GZA BO 13 ORHIONMWON ABD ED 25 OWERRI-MUNICIPAL WER IM 37 TARAUNI TRN KN 3 EDU LAF KW 14 IPOKIA PKA OG PLATEAU 3 DAMATURU DTR YB 6 ARM Pension Managers Limited 0026 S/N LGA NAME LGA STATE 12 HAWUL HWL BO 14 OVIA-NORTH-EAST AKA ED 26 26 OWERRI-NORTH RRT IM 38 TOFA TEA KN 4 EKITI ARP KW 15 OBAFEMI OWODE WDE OG S/N LGA NAME LGA STATE 4 FIKA FKA YB 7 Trustfund Pensions Plc 0028 CODE CODE 13 JERE JRE BO 15 OVIA-SOUTH-WEST GBZ ED 27 27 OWERRI-WEST UMG IM 39 TSANYAWA TYW KN 5 IFELODUN SHA KW 16 ODEDAH DED OG CODE CODE 5 FUNE FUN YB 8 First Guarantee Pension Limited 0029 1 AGUATA AGU AN 14 KAGA KGG BO 16 OWAN-EAST -

Analysis of Dynamical Rain Duration and Return Periods for Terrestrial and Satellite Communication Applications in a Tropical Climate
International Journal of Scientific & Engineering Research, Volume 7, Issue 2, February-2016 1305 ISSN 2229-5518 Analysis of dynamical rain duration and return periods for terrestrial and satellite communication applications in a tropical climate. Joseph Sunday Ojo Abstract— Understanding the intensity, duration and return periods of tropical rain events are critical to radio and satellite system engineers. As current and future satellite systems use and will use higher frequency bands, rain attenuation continues to pose more threat to link availability for microwave propagation operating in the frequencies above 10 GHz. Despite the enormous research on rain mitigations, few studies have addressed the dynamic of rain duration and return periods of tropical rain events. In this paper, an attempt has been made to analyze the dynamical rain duration and return periods for terrestrial and satellite communication applications in a tropical climate. 3-year rain rate data collected at Akure (7.17 oN, 5.18 oE) between July 2012 and June, 2015 is used. The dependence of integration time on the cumulative distribution of rain rate has been compared with results earlier obtained in Nigeria and other parts of the world. The Result shows that the power law relationship exists between the equiprobable rain rates of two different integration times. The value of conversion factors C E(R) and CR (R 0.01%) obtained for Akure are 0.15(30) and 0.33(110) respectively. The overall results show that different conversion factors are required for different locations, -

Kulikuli” and “Donkwa” in Ogbomoso and Ilorin, Nigeria
ACTA SCIENTIFIC NUTRITIONAL HEALTH (ISSN:2582-1423) Volume 4 Issue 4 April 2020 Research Article Microbiological Assessment of Selected Groundnut Based Snack “Kulikuli” and “Donkwa” in Ogbomoso And Ilorin, Nigeria 1 2 1 3 Adeoye AO , Adeyinka OO , Ajala AS * and Ojo MA Received: February 27, 2020 1 Department of Food Science and Engineering, Ladoke Akintola University of Published: March 14, 2020 Technology, Ogbomoso, Oyo State, Nigeria © All rights are reserved by Ajala AS., et al. 2Anatomy Department, Ladoke Akintola University of Technology, Ogbomoso, Oyo State, Nigeria 3Department of Food Science and Engineering, Ladoke Akintola University of Technology, Ogbomoso, Oyo State, Nigeria *Corresponding Author: Ajala AS, Department of Food Science and Engineering, Ladoke Akintola University of Technology, Ogbomoso, Oyo State, Nigeria. Abstract In this study, the microbial quality assessments of two peanut based snacks were analyzed. Samples were obtained from ‘Orita Naira’ market, Ogbomosho, Oyo State and ‘Oja Titun’ Ilorin, Kwara State, Nigeria. The controls were prepared under hygienic condi- tion in the Food processing laboratory, Ladoke Akintola University of Technology, Ogbomoso. These samples were analyzed to inves- tigate their safety. Thus, microbial colony estimation and characterization were carried out; these include microbial load, frequency of occurrence and antimicrobial sensitivity test. All the samples had bacterial and fungal contamination at varying degree; microbial load in terms of percentage frequency of occurrence of bacteria was between 5 - 30%, the percentage frequency of occurrence for fungi was 5 - 16%. For bacteria, the highest percentage microbial count was Salmonella spp (30%) while the least was E. coli (5%), while for fungi, the highest and lowest percentage microbial count were 16 and 5% respectively.