What Is Behind Hyperpermeability in Bradykinin-Mediated Angioedema?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

Binding Mode Exploration of B1 Receptor Antagonists' by the Use of Molecular Dynamics and Docking Simulation—How Different T
International Journal of Molecular Sciences Article Binding Mode Exploration of B1 Receptor Antagonists’ by the Use of Molecular Dynamics and Docking Simulation—How Different Target Engagement Can Determine Different Biological Effects Marica Gemei 1,*, Carmine Talarico 1 , Laura Brandolini 1, Candida Manelfi 1, Lorena Za 2, Silvia Bovolenta 2, Chiara Liberati 2, Luigi Del Vecchio 3, Roberto Russo 4 , Carmen Cerchia 4, Marcello Allegretti 1 and Andrea Rosario Beccari 1 1 Dompé Farmaceutici SpA, via Campo di Pile, 67100 L’Aquila, Italy; [email protected] (C.T.); [email protected] (L.B.); candida.manelfi@dompe.com (C.M.); [email protected] (M.A.); [email protected] (A.R.B.) 2 Axxam, Via Meucci 3, Bresso, 20091 Milano, Italy; [email protected] (L.Z.); [email protected] (S.B.); [email protected] (C.L.) 3 Ceinge Biotecnologie Avanzate, via G. Salvatore 486, 80145 Napoli, Italy; [email protected] 4 Department of Pharmacy, University of Naples “Federico II”, via D. Montesano, 49, 80131 Napoli, Italy; [email protected] (R.R.); [email protected] (C.C.) * Correspondence: [email protected]; Tel.: +34-06-465916 Received: 26 August 2020; Accepted: 12 October 2020; Published: 16 October 2020 Abstract: The kinin B1 receptor plays a critical role in the chronic phase of pain and inflammation. The development of B1 antagonists peaked in recent years but almost all promising molecules failed in clinical trials. Little is known about these molecules’ mechanisms of action and additional information will be necessary to exploit the potential of the B1 receptor. -

Quantigene Flowrna Probe Sets Currently Available
QuantiGene FlowRNA Probe Sets Currently Available Accession No. Species Symbol Gene Name Catalog No. NM_003452 Human ZNF189 zinc finger protein 189 VA1-10009 NM_000057 Human BLM Bloom syndrome VA1-10010 NM_005269 Human GLI glioma-associated oncogene homolog (zinc finger protein) VA1-10011 NM_002614 Human PDZK1 PDZ domain containing 1 VA1-10015 NM_003225 Human TFF1 Trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) VA1-10016 NM_002276 Human KRT19 keratin 19 VA1-10022 NM_002659 Human PLAUR plasminogen activator, urokinase receptor VA1-10025 NM_017669 Human ERCC6L excision repair cross-complementing rodent repair deficiency, complementation group 6-like VA1-10029 NM_017699 Human SIDT1 SID1 transmembrane family, member 1 VA1-10032 NM_000077 Human CDKN2A cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) VA1-10040 NM_003150 Human STAT3 signal transducer and activator of transcripton 3 (acute-phase response factor) VA1-10046 NM_004707 Human ATG12 ATG12 autophagy related 12 homolog (S. cerevisiae) VA1-10047 NM_000737 Human CGB chorionic gonadotropin, beta polypeptide VA1-10048 NM_001017420 Human ESCO2 establishment of cohesion 1 homolog 2 (S. cerevisiae) VA1-10050 NM_197978 Human HEMGN hemogen VA1-10051 NM_001738 Human CA1 Carbonic anhydrase I VA1-10052 NM_000184 Human HBG2 Hemoglobin, gamma G VA1-10053 NM_005330 Human HBE1 Hemoglobin, epsilon 1 VA1-10054 NR_003367 Human PVT1 Pvt1 oncogene homolog (mouse) VA1-10061 NM_000454 Human SOD1 Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

RT² Profiler PCR Array (Rotor-Gene® Format) Rat Pain: Neuropathic & Inflammatory
RT² Profiler PCR Array (Rotor-Gene® Format) Rat Pain: Neuropathic & Inflammatory Cat. no. 330231 PARN-162ZR For pathway expression analysis Format For use with the following real-time cyclers RT² Profiler PCR Array, Rotor-Gene Q, other Rotor-Gene cyclers Format R Description The Rat Pain: Neuropathic & Inflammatory RT² Profiler PCR Array profiles the expression of 84 genes involved in the transduction, maintenance, and modulation of pain responses. Noxious environmental stimuli, tissue damage, and disease all evoke pain. Since it afflicts up to 20% of the population at any given time, pain provides both a massive therapeutic target and a route to understanding the molecular mechanisms of nervous system function. While neuropathic pain often results from damage to the peripheral (PNS) or central nervous system (CNS), peripheral tissue damage and/or inflammation generally initiates inflammatory pain. Neuropathic and inflammatory pain both cause activation of damage-sensing neurons (nociceptors) that innervate the skin, muscle and viscera and terminate in the laminae of the spinal cord dorsal horn. Nociceptors conduct information to the CNS via neurotransmission and action potentials generated by ion channel and purinergic, opioid, and cannabinoid receptors leading to second order neuron activation. Synaptic transmission via glutamate, serotonin, and dopamine systems then follows. The transduction by nociceptors can be modulated by mediators of inflammation released by infiltrating immune cells and damaged neurons. Excitability of spinal neurons is also modulated by activation of resident microglia that release growth factors (such as BDNF), chemokines, and cytokines. Endogenous opioid peptides and arachidonic acid metabolites acting through G-protein coupled receptors also modulate neuronal excitability. -

Pharmacology of Basimglurant (RO4917523, RG7090), a Unique Mglu5 Negative Allosteric Modulator in Clinical Development for Depression
JPET Fast Forward. Published on February 9, 2015 as DOI: 10.1124/jpet.114.222463 This article has not been copyedited and formatted. The final version may differ from this version. JPET #222463 Title page Pharmacology of basimglurant (RO4917523, RG7090), a unique mGlu5 negative allosteric modulator in clinical development for depression Lothar Lindemann*, Richard H. Porter, Sebastian H. Scharf, Basil Kuennecke, Andreas Bruns, Markus von Downloaded from Kienlin, Anthony C. Harrison, Axel Paehler, Christoph Funk, Andreas Gloge, Manfred Schneider, Neil J. Parrott, Liudmila Polonchuk, Urs Niederhauser, Stephen R. Morairty, Thomas S. Kilduff, Eric Vieira, Sabine jpet.aspetjournals.org Kolczewski, Juergen Wichmann, Thomas Hartung, Michael Honer, Edilio Borroni, Jean-Luc Moreau, Eric Prinssen, Will Spooren, Joseph G. Wettstein, Georg Jaeschke at ASPET Journals on September 27, 2021 Roche Pharmaceutical Research and Early Development, Discovery Neuroscience, Neuroscience, Ophthalmology, and Rare Diseases (NORD) (L.L., S.H.S., B.K., A.B., M.v.K., M.H., E.B., E.P., W.S., J.G.W.), Discovery Chemistry (E.V., S.K., J.W., G.J.), Operations for Neuroscience, Ophthalmology, and Rare Diseases (R.H.P., J.-L.M.), Pharmaceutical Sciences (A.H., A.P., C.F., A.G., M.S., N.P., L.P., U.N.), Small Molecules Process Research and Synthesis (T.H.), Roche Innovation Center Basel, Grenzacherstrasse 124, CH-4070 Basel, Switzerland Center for Neuroscience, Biosciences Division, SRI International, Menlo Park, California 94025, USA (S.R.M., T.S.K.) 1 JPET Fast Forward. Published on February 9, 2015 as DOI: 10.1124/jpet.114.222463 This article has not been copyedited and formatted. -

Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 12-2013 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Medical Cell Biology Commons, and the Medical Molecular Biology Commons Recommended Citation Bogard, Amy Sue , "Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells" (2013). Theses and Dissertations (ETD). Paper 330. http://dx.doi.org/10.21007/etd.cghs.2013.0029. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Document Type Dissertation Degree Name Doctor of Philosophy (PhD) Program Biomedical Sciences Track Molecular Therapeutics and Cell Signaling Research Advisor Rennolds Ostrom, Ph.D. Committee Elizabeth Fitzpatrick, Ph.D. Edwards Park, Ph.D. Steven Tavalin, Ph.D. Christopher Waters, Ph.D. DOI 10.21007/etd.cghs.2013.0029 Comments Six month embargo expired June 2014 This dissertation is available at UTHSC Digital Commons: https://dc.uthsc.edu/dissertations/330 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells A Dissertation Presented for The Graduate Studies Council The University of Tennessee Health Science Center In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy From The University of Tennessee By Amy Sue Bogard December 2013 Copyright © 2013 by Amy Sue Bogard. -

Bradykinin Pathway and Pathological States: Potential Therapeutic Agents Against Such States
Editorial American Journal of Pharmacology Published: 16 Feb, 2021 Bradykinin Pathway and Pathological States: Potential Therapeutic Agents against Such States Chika J Mbah* Department of Pharmaceutical and Medicinal Chemistry, University of Nigeria, Nigeria Editorial Bradykinin is one of the two kinins that are present in human body. It is a biologically active nonapeptide hormone released following the breakdown of kallidin (decapeptide lysyl-bradykinin) by aminopeptidase [1]. Kallidin is generated from low molecular weight kininogen by the action of kininogenases. Kininogenases include plasma kallikrein and tissue kallikrein. Kininogen is found in the α2-globulin fraction of normal mammalian plasma [2]. Bradykinin is very effectively removed during passage through the lung and other vascular beds. It is degraded by peptidases. The cellular effects of bradykinin are mediated by two bradykinin G-protein-coupled receptors namely Bradykinin Receptor B1 (B1BKR) and Bradykinin Receptor B2 (B2BKR) respectively [3]. Bradykinin receptor B2 is constitutively expressed in numerous tissues and after ligand binding becomes promptly desensitized. The low expression of bradykinin receptor B1 in healthy conditions however becomes increased during inflammation, infection, or injury [4,5]. The mechanism of action may involve (i) stimulation of the membrane phospholipid metabolism by activating phospholipase C, (ii) promotion of intracellular calcium mobilization by inositol 1,4,5-triphosphate, (iii) release of Nitric Oxide (NO) and prostaglandins [4]. The disruption or loss of homeostasis associated with bradykinin activation may result in disease states such as (i) inflammation-arising from vasodilation, increased vascular permeability, stimulating prostaglandins synthesis [6] (ii) spinal cord injury [7] (iii) central and peripheral nervous systems ischemia [8] (iv) plasma extravasation [9] (v) bronchoconstriction-linked to asthma and rhinitis [10] (vi) pain and irritation in skin, muscle, joints, vasculature, and all visceral organs [11,12]. -
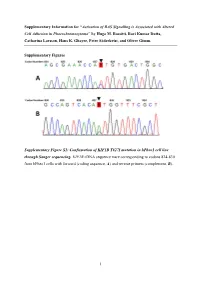
Activation of RAS Signalling Is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M
Supplementary Information for “Activation of RAS Signalling is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M. Rossitti, Ravi Kumar Dutta, Catharina Larsson, Hans K. Ghayee, Peter Söderkvist, and Oliver Gimm. Supplementary Figures Supplementary Figure S1: Confirmation of KIF1B T827I mutation in hPheo1 cell line through Sanger sequencing. KIF1B cDNA sequence trace corresponding to codons 824-830 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 1 Supplementary Figure S2: Confirmation of NRAS Q61K mutation in hPheo1 cell line through Sanger sequencing. NRAS cDNA sequence trace corresponding to codons 56-67 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 2 Supplementary Figure S3: CCND1 gene expression and hPheo1 proliferation. A: Expression of CCND1 mRNA assessed by RT-qPCR and presented as fold change (2-ΔΔCT, mean ± standard error of the mean). B: Cell counts at 1, 2, and 3 days after plating (corresponding to 72, 96 and 120 hours posttransfection, respectively) of control- or siNRAS#1-transfected hPheo1 cells expressed as fold change of the number of cells plated at day 0 (48 hours posttransfection; mean ± standard deviation). All results are from three independent siRNA experiments. 3 Supplementary Tables Supplementary Table S1: List of transcript cluster IDs significantly upregulated in hPheo1 by siNRAS treatment (comparison: siNRAS versus control-transfected hPheo1; ANOVA p < 0.05, FDR < 0.25, fold change < -1.5 or > 1.5). Transcript -
List of Predicted Circfam120a Target Mrnas Gene in Pathway Species Gene Name
List of predicted circFAM120A target mRNAs Gene in pathway Species Gene name HRAS Homo sapiens HRas proto-oncogene, GTPase (HRAS) ADCY1 Homo sapiens Adenylate cyclase 1 (ADCY1) ADCY2 Homo sapiens Adenylate cyclase 2 (ADCY2) ADCY7 Homo sapiens Adenylate cyclase 7 (ADCY7) PTGS2 Homo sapiens Prostaglandin-endoperoxide synthase 2 (PTGS2) PGF Homo sapiens Placental growth factor (PGF) ADCY5 Homo sapiens Adenylate cyclase 5 (ADCY5) STAT5A Homo sapiens Signal transducer and activator of transcription 5A (STAT5A) ADCY6 Homo sapiens Adenylate cyclase 6 (ADCY6) STAT5B Homo sapiens Signal transducer and activator of transcription 5B (STAT5B) LPAR3 Homo sapiens Lysophosphatidic acid receptor 3 (LPAR3) LPAR2 Homo sapiens Lysophosphatidic acid receptor 2 (LPAR2) CTNNB1 Homo sapiens Catenin beta 1 (CTNNB1) CUL2 Homo sapiens Cullin 2 (CUL2) RARA Homo sapiens Retinoic acid receptor alpha (RARA) GNG2 Homo sapiens G protein subunit gamma 2 (GNG2) RARB Homo sapiens Retinoic acid receptor beta (RARB) GNG3 Homo sapiens G protein subunit gamma 3 (GNG3) FAS Homo sapiens Fas cell surface death receptor (FAS) GNG4 Homo sapiens G protein subunit gamma 4 (GNG4) GNG7 Homo sapiens G protein subunit gamma 7 (GNG7) PIK3CG Homo sapiens Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma (PIK3CG) WNT10B Homo sapiens Wnt family member 10B (WNT10B) BCR Homo sapiens BCR, RhoGEF and GTPase activating protein (BCR) BRAF Homo sapiens B-Raf proto-oncogene, serine/threonine kinase (BRAF) RXRB Homo sapiens Retinoid X receptor beta (RXRB) ROCK2 Homo sapiens Rho -

Regulation Pathway of Mesenchymal Stem Cell Immune Dendritic Cell
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2010; 185:5102-5110; Prepublished online 1 from submission to initial decision 4 weeks from acceptance to publication October 2010; doi: 10.4049/jimmunol.1001332 http://www.jimmunol.org/content/185/9/5102 Inhibition of Immune Synapse by Altered Dendritic Cell Actin Distribution: A New Pathway of Mesenchymal Stem Cell Immune Regulation Alessandra Aldinucci, Lisa Rizzetto, Laura Pieri, Daniele Nosi, Paolo Romagnoli, Tiziana Biagioli, Benedetta Mazzanti, Riccardo Saccardi, Luca Beltrame, Luca Massacesi, Duccio Cavalieri and Clara Ballerini J Immunol cites 38 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2010/10/01/jimmunol.100133 2.DC1 This article http://www.jimmunol.org/content/185/9/5102.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2010 by The American Association of -

Mrna Expression in Human Leiomyoma and Eker Rats As Measured by Microarray Analysis
Table 3S: mRNA Expression in Human Leiomyoma and Eker Rats as Measured by Microarray Analysis Human_avg Rat_avg_ PENG_ Entrez. Human_ log2_ log2_ RAPAMYCIN Gene.Symbol Gene.ID Gene Description avg_tstat Human_FDR foldChange Rat_avg_tstat Rat_FDR foldChange _DN A1BG 1 alpha-1-B glycoprotein 4.982 9.52E-05 0.68 -0.8346 0.4639 -0.38 A1CF 29974 APOBEC1 complementation factor -0.08024 0.9541 -0.02 0.9141 0.421 0.10 A2BP1 54715 ataxin 2-binding protein 1 2.811 0.01093 0.65 0.07114 0.954 -0.01 A2LD1 87769 AIG2-like domain 1 -0.3033 0.8056 -0.09 -3.365 0.005704 -0.42 A2M 2 alpha-2-macroglobulin -0.8113 0.4691 -0.03 6.02 0 1.75 A4GALT 53947 alpha 1,4-galactosyltransferase 0.4383 0.7128 0.11 6.304 0 2.30 AACS 65985 acetoacetyl-CoA synthetase 0.3595 0.7664 0.03 3.534 0.00388 0.38 AADAC 13 arylacetamide deacetylase (esterase) 0.569 0.6216 0.16 0.005588 0.9968 0.00 AADAT 51166 aminoadipate aminotransferase -0.9577 0.3876 -0.11 0.8123 0.4752 0.24 AAK1 22848 AP2 associated kinase 1 -1.261 0.2505 -0.25 0.8232 0.4689 0.12 AAMP 14 angio-associated, migratory cell protein 0.873 0.4351 0.07 1.656 0.1476 0.06 AANAT 15 arylalkylamine N-acetyltransferase -0.3998 0.7394 -0.08 0.8486 0.456 0.18 AARS 16 alanyl-tRNA synthetase 5.517 0 0.34 8.616 0 0.69 AARS2 57505 alanyl-tRNA synthetase 2, mitochondrial (putative) 1.701 0.1158 0.35 0.5011 0.6622 0.07 AARSD1 80755 alanyl-tRNA synthetase domain containing 1 4.403 9.52E-05 0.52 1.279 0.2609 0.13 AASDH 132949 aminoadipate-semialdehyde dehydrogenase -0.8921 0.4247 -0.12 -2.564 0.02993 -0.32 AASDHPPT 60496 aminoadipate-semialdehyde