New World Cattle Show Ancestry from Multiple Independent Domestication Events
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

VII: Sire Types for Commercial Herds
E-191 7-03 VII: Sire Types for Commercial Herds for Beef Cattle Stephen P. Hammack* Choosing types of sires is one of the most important genetic deci- Producers who document and merchandise true genetic merit or, sions facing beef producers. particularly, retain ownership can be most flexible in choosing breed That choice depends on: types. Those who market through traditional methods are subject to biases and perceptions, often resulting in price differences that may Climatic and management conditions and number of production phases involved not be justified but are nevertheless real. These traditional producers can maximize production efficiency and avoid or minimize severe Breeding systems used price discounts by producing medium- to large-frame crossbred Breeds or types and individual performance levels calves of at least 1/4 British, no more than 1/2 Continental, no more Types of cows currently in the herd. than 1/4 Bos indicus and no more than 1/4 Dairy. For high-quality mar- Production conditions must be assessed accurately to avoid kets, higher percentages of British are desirable. For lean-beef mar- incompatibility caused by too much or too little genetic production kets, higher percentages of Continental are applicable. potential. For a discussion of two important genetic factors, see E- Some price difference exists even within the above ranges that: 188, “Texas Adapted Genetic Strategies for Beef Cattle—III: Body Varies over time as to the exact breed-type percentages Size and Milking Level.” Also, genetic considerations may not be the favored same for marketing at weaning as for retained ownership, especially Is usually small and short-term, compared to differences for when selling on a carcass grid. -

Multiple Choice Choose the Answer That Best Completes Each Statement Or Question
Name Date Hour 5 The Beef Cattle Industry Multiple Choice Choose the answer that best completes each statement or question. _______ 1. Early settlers primarily used cattle as ____ . A. work animals B. a source of meat C. a source of milk D. symbols of wealth _______ 2. The modern cattle industry is concentrated in the ____ . A. South and Midwest B. South and Southwest C. North and Northwest D. North and Midwest _______ 3. Cattle drives were necessary in the past because of the lack of ____ . A. retail stores B. refrigeration C. slaughterhouses D. year-round grazing _______ 4. Which beef cattle breed is known for its solid black color and excellent meat quality? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 5. Which beef cattle breed is from northern England and was often called a Durham after the county in which it originated? A. Angus B. Hereford C. Shorthorn D. Chianina Introduction to Agriscience | Unit 5 Test CIMC 1 _______ 6. Which beef cattle breed originated in England and was originally much larger, weighing more than 3,000 pounds, than it is today? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 7. Which beef cattle breed is red with a white face and may also have white on the neck, underline, legs, and tail switch? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 8. Which beef cattle breed is one of the oldest breeds in the world and originated in Italy? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 9. Which beef cattle breed originated in central France and was developed as a dual-purpose breed and is typically white or off-white in color? A. -
Proc1-Beginning Chapters.Pmd
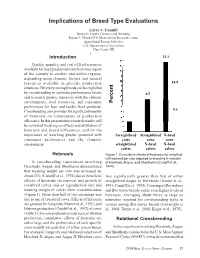
Implications of Breed Type Evaluations Larry V. Cundiff Research Leader, Genetics and Breeding Roman L. Hruska U.S. Meat Animal Research Center Agricultural Research Service U.S. Department of Agriculture Clay Center, NE ()(,) Introduction 23.3 Quality, quantity, and cost of feed resources available for beef production vary from one region of the country to another and within regions, depending upon climatic factors and natural 23.3 resources available in specific production 14.8 situations. Diversity among breeds can be exploited by crossbreeding to optimize performance levels 8.5 and to match genetic resources with the climatic environment, feed resources, and consumer Percent preferences for lean and tender beef products. 8.5 Crossbreeding also provides for significant benefits 14.8 of heterosis on components of production efficiency. In this presentation, research results will be reviewed focusing on effects and utilization of 8.5 8.5 heterosis and breed differences, and on the importance of matching genetic potential with Straightbred Straightbred X-bred consumer preferences and the climatic cows cows cows environment. straightbred X-bred X-bred calves calves calves Heterosis Figure 1. Cumulative effects of heterosis for weight of calf weaned per cow exposed to breeding in crosses A crossbreeding experiment involving of Hereford, Angus, and Shorthorns (Cundiff et al., Herefords, Angus, and Shorthorns demonstrated 1974). that weaning weight per cow was increased by about 23% (Cundiff et al., 1974) due to beneficial was significantly greater than that of either effects of heterosis on survival and growth of straightbred Angus or Herefords (Nunez et al., crossbred calves and on reproduction rate and 1991; Cundiff et al., 1992). -

Comparing SNP Panels and Statistical Methods for Estimating Genomic Breed Composition of Individual Animals in Ten Cattle Breeds
He et al. BMC Genetics (2018) 19:56 https://doi.org/10.1186/s12863-018-0654-3 RESEARCH ARTICLE Open Access Comparing SNP panels and statistical methods for estimating genomic breed composition of individual animals in ten cattle breeds Jun He1,2†,YageGuo1,3†, Jiaqi Xu1,4, Hao Li1,5,AnnaFuller1, Richard G. Tait Jr1,Xiao-LinWu1,5* and Stewart Bauck1 Abstract Background: SNPs are informative to estimate genomic breed composition (GBC) of individual animals, but selected SNPs for this purpose were not made available in the commercial bovine SNP chips prior to the present study. The primary objective of the present study was to select five common SNP panels for estimating GBC of individual animals initially involving 10 cattle breeds (two dairy breeds and eight beef breeds). The performance of the five common SNP panels was evaluated based on admixture model and linear regression model, respectively. Finally, the downstream implication of GBC on genomic prediction accuracies was investigated and discussed in a Santa Gertrudis cattle population. Results: There were 15,708 common SNPs across five currently-available commercial bovine SNP chips. From this set, four subsets (1,000, 3,000, 5,000, and 10,000 SNPs) were selected by maximizing average Euclidean distance (AED) of SNP allelic frequencies among the ten cattle breeds. For 198 animals presented as Akaushi, estimated GBC of the Akaushi breed (GBCA) based on the admixture model agreed very well among the five SNP panels, identifying 166 animals with GBCA = 1. Using the same SNP panels, the linear regression approach reported fewer animals with GBCA = 1. -

DOMESTIC CATTLE ARTIODACTYLA Family: Bovidae Genus: Bos Species: Taurus
DOMESTIC CATTLE ARTIODACTYLA Family: Bovidae Genus: Bos Species: taurus Range: world wide Habitat: open grass lands, rangelands Miniature Corriente Cattle Niche: terrestrial, diurnal, herbivorous Wild diet: grasses, stems Zoo diet: Life Span: 25 years Sexual dimorphism: M larger than F Location in SF Zoo: Family Farm in the Exploration Zone APPEARANCE & PHYSICAL ADAPTATIONS: Cattle are large ungulates with cloven hooves; the hoof is split into two toes which are homologous to the third and fourth digits. Most breeds have horns, which vary in size among the breeds. Genetic selection has allowed polled (hornless) cattle to become widespread. Weight: ~ 1,660 lbs varies among breeds HRL: Domestic cows have no upper incisors, instead they have a thick SH: 49 – 52 in layer called the dental pad. The jaws are designed for the circular TL: grinding motion used to crush coarse vegetation. Cattle are ruminants or “cud chewers”, with four-chambered stomach and foregut fermentation by microbes; they are highly specialized to eat poorly digestible plants as food. These microbes are primarily responsible for decomposing the cellulose of the herbivorous diet. The microbes reproduce in the rumen, older generations die and their cells continue on through the digestive tract. These cells are then partially digested in the small intestines, allowing cattle to gain a high-quality protein source. Cattle are dichromatic and having two kinds of color receptors in their retinas, as are most other non-primate land mammals. STATUS & CONSERVATION Domestic cows are common and can be found throughout the world. A report from the Food and Agriculture Organization (FAO) states that the livestock sector is "responsible for 18% of greenhouse gas emissions". -

The Texas Longhorn the First Long-Horned Cattle Came to North America Between the Sixteenth and Eighteenth Centuries During
The Texas Longhorn The first long-horned cattle came to North America between the sixteenth and eighteenth centuries during the Spanish explorations, expeditions and religious missions. When Anglo-American pioneers traveled west in the early nineteenth century, they brought domesticated English cattle with them. American Indians raided cattle from both the Spanish and Anglo-Americans and gradually developed their own hybrid strains of cattle. With many cattle escaping from the open ranges, Spanish, English and hybrid cattle interbred. By the 1830’s, thousands of wild cattle ranged from the Rio Grande to the Nueces River. Exposed to the elements and relentlessly hunted by American Indians and colonists, cattle adapted or perished. They were forced to survive in the blazing sun and freezing winter, through dust storms and swamps. They could subsist on a diet of weeds and brush and live for days without water. From their Spanish ancestors, they inherited large, sweeping horns with twists at the end that allowed them to ward off coyotes and wolves. Colonists called cattle of the brush “wild cattle,” “mustang cattle” or “Spanish cattle.” It was not until the end of the Civil War that some range men would refer to “Texas cattle” or “Texas Longhorns.” Early Texas cowboys rustled cattle from Mexican ranches and captured feral cattle from the brush country. They stocked Texas ranges with these animals or trailed them through the Louisiana swamps to markets in New Orleans. Although the animals varied in their degree of wildness, skilled raiders could drive them in herds. After two or three long days of running, then trotting, then walking, they could be managed almost like domestic cattle. -

Purebred Livestock Registry Associations
Purebred livestock registry associations W. Dennis Lamm1 COLORADO STATE UNIVERSITY EXTENSION SERVICE no. 1.217 Beef Devon. Devon Cattle Assn., Inc., P.O. Box 628, Uvalde, TX 78801. Mrs. Cammille Hoyt, Sec. Phone: American. American Breed Assn., Inc., 306 512-278-2201. South Ave. A, Portales, NM 88130. Mrs. Jewell Dexter. American Dexter Cattle Assn., P.O. Jones, Sec. Phone: 505-356-8019. Box 56, Decorah, IA 52l01. Mrs. Daisy Moore, Amerifax. Amerifax Cattle Assn., Box 149, Exec. Sec. Phone: 319-736-5772, Hastings, NE 68901. John Quirk, Pres. Phone Friesian. Beef Friesian Society, 213 Livestock 402-463-5289. Exchange Bldg., Denver, CO 80216. Maurice W. Angus. American Angus Assn., 3201 Freder- Boney, Adm. Dir. Phone: 303-587-2252. ick Blvd., St. Joseph, MO 64501. Richard Spader, Galloway. American Galloway Breeders Assn., Exec. Vice. Pres. Phone: 816-233-3101. 302 Livestock Exchange Bldg., Denver, CO 80216. Ankina. Ankina Breeders, Inc., 5803 Oaks Rd,. Cecil Harmon, Pres. Phone: 303-534-0853. Clayton, OH 45315. James K. Davis, Ph.D., Pres. Galloway. Galloway Cattle Society of Amer- Phone: 513-837-4128. ica, RFD 1, Springville, IA 52336. Phone: 319- Barzona. Barzona Breeders Assn. of America, 854-7062. P.O. Box 631, Prescott, AZ 86320. Karen Halford, Gelbvieh. American Gelbvieh Assn., 5001 Na- Sec. Phone: 602-445-2290. tional Western Dr., Denver, CO 80218. Daryl W. Beefalo. American Beefalo Breeders, 1661 E. Loeppke, Exec. Dir. Phone: 303-296-9257. Brown Rd., Mayville 22, MI 48744. Phone: 517-843- Hays Convertor. Canadian Hays Convertor 6811. Assn., 6707 Elbow Dr. SW, Suite 509, Calgary, Beefmaster. -

Florida Agriculture Statistical Directory
Dear Friends of Agriculture, It is my pleasure to present the 2008 Florida Agriculture Statistical Directory. This report presents a wealth of information about Florida’s vast and varied agricultural production through data that details land use, crop yields, commodity prices, crop rankings and more. This yearly report is invaluable to anyone who is involved in this dynamic business or who wants to better understand its complexities. The tables, charts and statistics contained in this report do an exceptional job of measuring the inputs and outputs, and presenting Florida agriculture in the context of “hard numbers.” But there is more to our state’s agricultural industry: our hard-working farmers, whose dedication, hard work and perseverance have made Florida agriculture into the diverse and highly productive industry that is respected throughout the globe. As evidenced by the ever-growing popularity of the “Fresh from Florida” label, consumers worldwide appreciate and seek out the quality products that our farmers provide. Maintaining these standards of excellence seldom comes easily as each year presents new challenges for Florida’s 40,000 commercial farmers. But, whether confronted by hurricanes, freezes, pests, diseases or fierce international competition, our state’s producers continually show that they are up to the test. Enterprising spirit, love of the land, and pride in their products are all hallmarks of the well- earned reputation of Florida’s farmers. In addition to enjoying the quality products that our farmers produce, Florida’s agricultural production benefits our state’s residents in other important ways as well. Florida agriculture has an overall economic impact estimated at more than $100 billion annually, making it a sound pillar of the state’s economy. -

Butler Breeder's Invitational 20Th Anniversary Sale
Schedule of Events BUTLER BREEDER’S INVITATIONAL 20TH ANNIVERSARY SALE Labor Day Weekend Saturday, September 2, 2017, 12:00 Noon 12:00 p.m. Pre-Sale Activities Cattle begin selling approximately 12:30 p.m. Lockhart Auction – Highway 183 S – Lockhart, Texas (28 miles south of Austin) Friday, September 1, 2017 12:00 Noon Cattle Viewing Saturday, September 2, 2017 8:00 a.m. Cattle Viewing 11:00 a.m. Lunch SALE HEADQUARTERS: SALE DAY PHONE: 325-473-1373 Lockhart Plum Creek Inn 512-398-4911 Auctioneer: Joel Lemley Best Western 512-620-0300 Pedigrees: Kaso Kety Additional hotel space conveniently Cattle Handler: Michael McLeod Located in Austin, Bastrop, Luling & San Marcos Ringmen: Wyman Poe Airports: Lockhart Municipal Airport Troy Robinett Bergstrom Airport – Austin Haulers: Cody James San Antonio International 903-695-2564 Home 903-519-4043 Cell LIVE WEBCAST & INTERNET BIDDING AVAILABLE LIVE WEBCAST begins with Pre-Sale Activities at 12:00 on SATURDAY www.CattleInMotion.com SPONSORED BY: Butler Breeder’s Invitational Sale To view the WEBCAST, go to www.CattleInMotion.com Butler Longhorn Museum Select BUTLER BREEDER’S INVITATIONAL from Upcoming Events. Falls Creek Ranch All viewers and bidders must create an account. Jane’s Land and Cattle Company OnLine Bidders must be pre-approved by September 1. Rio Vista Ranch/Rocking P Longhorns Follow instructions listed on the Cattle In Motion website. Phone Bids may be placed by calling the Sale Day Phone: 325-473-1373 Phone Bidding must be pre-approved by August 31, 2017 For Pre-approval, call: 325-668-3552 or 325-473-1373 – 1 – Consignors BPT Longhorns Kety/Sellers Partnership Potts/Rosenberger Stanley Cattle Co. -

ACE Appendix
CBP and Trade Automated Interface Requirements Appendix: PGA August 13, 2021 Pub # 0875-0419 Contents Table of Changes .................................................................................................................................................... 4 PG01 – Agency Program Codes ........................................................................................................................... 18 PG01 – Government Agency Processing Codes ................................................................................................... 22 PG01 – Electronic Image Submitted Codes .......................................................................................................... 26 PG01 – Globally Unique Product Identification Code Qualifiers ........................................................................ 26 PG01 – Correction Indicators* ............................................................................................................................. 26 PG02 – Product Code Qualifiers ........................................................................................................................... 28 PG04 – Units of Measure ...................................................................................................................................... 30 PG05 – Scientific Species Code ........................................................................................................................... 31 PG05 – FWS Wildlife Description Codes ........................................................................................................... -

Each New GGP Generation Gives You More Advantages
A New Generation The GeneSeek® Genomic Profiler™ (GGP) portfolio empowers your selection, management and marketing of beef seedstock Each new GGP generation gives you more advantages Our GeneSeek Genomic Profiler (GGP) products and services empower your decisions in selecting, raising and selling elite cattle, enhancing profit and protecting your reputation for high-quality seedstock. Beef genomics is evolving fast. Neogen gives you the most advanced, widest range of DNA testing for the real world of cattle production. Why Neogen? • From partnering with all major breeds…to the industry’s broadest line of genomic profilers • From faster, easier DNA sampling at chute side…to world-class achievements like genotyping embryos • From running millions of DNA samples…to driving down the cost of genomic testing • From advanced data pipelines…to ongoing discovery on the genomic frontier • From field support for your operation…to global collaborations with world-renowned scientists • From defect and condition screening…to customized profiles for your breed If you want to be generations ahead, go with GGP. How to obtain GGP products and services GGP products are available through partner breed associations and genetics companies. GGP data are used to genomically enhance Expected Progeny Differences (GE-EPDs), test parentage and screen for genetic conditions. GGP data are transmitted to partners via secure, industry-leading bio-informatic tools and backed by our data experts and quality control team. October 1990 March 1998 July 2001 July 2001 Human Genome Project begins a National Beef Cattle A Timeline of Abe Oommen and Daniel Pomp Jim Gibb launches 13-year, $2.7 billion effort, world's Evaluation Consortium funded to found GeneSeek Frontier Beef Systems Genomic Milestones largest research collaboration improve beef genetics 2 The generational timelines of GE-EPDs & traditional bull test evaluation Collecting good quality phenotype data is very important. -

American Registered Breeds (Arb) Other Registered
AMERICAN REGISTERED BREEDS (ARB) 1. Open to heifers of Bos indicus type that have been registered or issued a certificate of recordation with a recognized breed association. The breeds that will be recognized are defined below: • Beefmaster Advancer - Animals of fifty percent (50%) or more Registered Beefmaster breeding and fifty percent (50%) or less of other Registered and DNA genotyped non-Beefmaster Beef cattle breeding. • Braford - Purebred, Heifers must be classified as Braford to be eligible to show. No F-1, multiple generation half blood, ¾ Hereford, ¾ Brahman, Single Bar or other percentage cattle will be accepted. • Brangus Premium Gold – Progeny of Registered Brangus or Red Brangus and any commercial animal. Must maintain a minimum of 50% registered Brangus/Red Brangus genetics. • Brangus Optimizer – Progeny resulting from mating Registered Brangus or Red Brangus and a registered animal from another beef breed recognized by the U.S. Beef Breeds Council, other than Angus or Red Angus. Must maintain a minimum of 50% registered Brangus genetics. • Brangus UltraBlack – Progeny of Registered Brangus and an Enrolled Angus • Brangus UltraRed- Progeny of Registered Red Brangus and an Enrolled Angus/Red Angus • Certified Beefmaster E6 - Certified by Beefmaster Breeders United to be at least 50% Beefmaster and can be as much as 100%. At least one of the parents must be registered as a purebred Beefmaster. • Golden Certified F1 – A female that is the progeny of two registered parents with one parent being registered Brahman resulting in a F1 cross (50% Brahman x 50% Bos Taurus). Must be issued a certificate of recordation form from ABBA which includes owner’s name, ownership date.