Comparative Study of Four Fluorinated Quinolones in Susceptibility Factors on Microorganisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Current Case of Quinolones: Synthetic Approaches and Antibacterial Activity
molecules Review The Current Case of Quinolones: Synthetic Approaches and Antibacterial Activity Abdul Naeem 1, Syed Lal Badshah 1,2,*, Mairman Muska 1, Nasir Ahmad 2 and Khalid Khan 2 1 National Center of Excellence in Physical Chemistry, University of Peshawar, Peshawar, Khyber Pukhtoonkhwa 25120, Pakistan; [email protected] (A.N.); [email protected] (M.M.) 2 Department of Chemistry, Islamia College University Peshawar, Peshawar, Khyber Pukhtoonkhwa 25120, Pakistan; [email protected] (N.A.); [email protected] (K.K.) * Correspondence: [email protected]; Tel.: +92-331-931-6672 Academic Editor: Peter J. Rutledge Received: 23 December 2015 ; Accepted: 15 February 2016 ; Published: 28 March 2016 Abstract: Quinolones are broad-spectrum synthetic antibacterial drugs first obtained during the synthesis of chloroquine. Nalidixic acid, the prototype of quinolones, first became available for clinical consumption in 1962 and was used mainly for urinary tract infections caused by Escherichia coli and other pathogenic Gram-negative bacteria. Recently, significant work has been carried out to synthesize novel quinolone analogues with enhanced activity and potential usage for the treatment of different bacterial diseases. These novel analogues are made by substitution at different sites—the variation at the C-6 and C-8 positions gives more effective drugs. Substitution of a fluorine atom at the C-6 position produces fluroquinolones, which account for a large proportion of the quinolones in clinical use. Among others, substitution of piperazine or methylpiperazine, pyrrolidinyl and piperidinyl rings also yields effective analogues. A total of twenty six analogues are reported in this review. The targets of quinolones are two bacterial enzymes of the class II topoisomerase family, namely gyrase and topoisomerase IV. -

Belgian Veterinary Surveillance of Antibacterial Consumption National
Belgian Veterinary Surveillance of Antibacterial Consumption National consumption report 2020 Publication : 22 June 2021 1 SUMMARY This annual BelVet-SAC report is now published for the 12th time and describes the antimicrobial use (AMU) in animals in Belgium in 2020 and the evolution since 2011. For the third year this report combines sales data (collected at the level of the wholesalers-distributors and the compound feed producers) and usage data (collected at farm level). This allows to dig deeper into AMU at species and farm level in Belgium. With a consumption of 87,6 mg antibacterial compounds/kg biomass an increase of +0.2% is seen in 2020 in comparison to 2019. The increase seen in 2020 is spread over both pharmaceuticals (+0.2%) and antibacterial premixes (+4.0%). This unfortunately marks the end of a successful reduction in antibacterial product sales that was seen over the last 6 years resulting in a cumulative reduction of -40,2% since 2011. The gap seen in the coverage of the sales data with the Sanitel-Med collected usage data increased substantially compared to 2019, meaning continuous efforts need to be taken to ensure completeness of the collected usage data. When looking at the evolution in the number of treatment days (BD100) at the species level, as calculated from the SANITEL- MED use data, use increased in poultry (+5,0%) and veal calves (+1,9%), while it decreased in pigs (-3,1%). However, the numerator data for this indicator remain to be updated for 2020, potentially influencing the reliability of the result. -

Development of an Antibiotic Policy in a Zoological Medicine Service and Approach to Antibiotic Dosing Using Minimum Inhibitory Concentration Data Stephen J
Development of an Antibiotic Policy in a Zoological Medicine Service and Approach to Antibiotic Dosing Using Minimum Inhibitory Concentration Data Stephen J. Divers, BVetMed, DZooMed, DipECZM (Herpetology), DipECZM (Zoo Health Management), Dipl ACZM, FRCVS, Izidora Sladakovic, BVSc (Hons I), MVS, Jörg Mayer, DVM, MS, DipECZM (Exotic Mammals), Dipl ACZM, DABVP (Exotic Companion Mammal Practice), Susan Sanchez, BSc, MSc, PhD, FRSB Session #2101 Affiliation: From The Department of Small Animal Medicine and Surgery (Divers, Sladakovic, Mayer) and Athens Veterinary Diagnostic Laboratory (Sanchez), College of Veterinary Medicine, University of Georgia, Athens, GA 30602, USA. Abstract: Antimicrobial resistance is a major veterinary concern, and yet despite widespread literature and policies advocating appropriate antibiotic stewardship in domesticated veterinary medicine, there appears to have been very little progress in zoological medicine, especially companion exotic practice. An example of an antibiotic policy for a zoological medicine service is described, along with general advice on appropriate antibiotic use, including minimum inhibitory concentration (MIC)-determined drug dosing. Introduction Antimicrobial resistance (AMR) is a global veterinary and public health challenge, which has accelerated by the overuse of antibiotics worldwide. Increased antimicrobial resistance is the cause of severe infections, complica- tions, longer hospital stays and increased mortality. Overprescribing of antibiotics is associated with an increased risk -

Supplementary Materials Comparison of Sample Preparations For
Supplementary Materials Comparison of sample preparations for determining the veterinary drug residues using liquid chromatography-tandem mass spectrometry in fishery products. Joohye Kima, Hyunjin Parka, Hui-Seung Kang*a a Pesticide and Veterinary Drug Residues Division, National Institute of Food and Drug Safety Evaluation, Osong, Chungcheongbuk-do 363-700, South Korea Table S1. Manufacturer of standards Compounds Manufacturer Compounds Manufacturer 4-MAP Albendazole Fluka (4-methylamino antipyrine) Sigma-Aldrich 2-Amino albendazole sulfone Dr. Ehrenstorfer Sarafloxacin Dr. Ehrenstorfer Albendazole sulfone Sigma-Aldrich Orbifloxacin Dr. Ehrenstorfer Albendazole sulfoxide Sigma-Aldrich Carbadox Sigma-Aldrich QCA Febantel Dr. Ehrenstorfer (Quinoxaline-2-carboxylic acid) Sigma-Aldrich Fenbendazole Sigma-Aldrich Olaquindox Wako MQCA Flubendazole Dr. Ehrenstorfer (3-methylquinoxaline-2- Sigma-Aldrich carboxylic acid) 2-amino flubendazole Sigma-Aldrich Dapsone Dr. Ehrenstorfer Oxfendazole Dr. Ehrenstorfer N-acethyl dapsone TRC Oxfendazole sulfone Wako Sulfapyridine Sigma-Aldrich Oxibendazole Sigma-Aldrich Arprinocid TRC Cefapirin Fluka Azaperol Sigma-Aldrich Desacetylcefapirin TRC Azaperon Fluka Cefazoline USP Carazolol Dr. Ehrenstorfer Cefoperazone USP Caffeine Sigma-Aldrich Halofuginone Sigma-Aldrich Clenbuterol Dr. Ehrenstorfer Azithromycin USP Colchicine Sigma-Aldrich Tildipirosin TRC Diphenhydramine Sigma-Aldrich Dimetridazole Dr. Ehrenstorfer Flunixin Dr. Ehrenstorfer Ipronidazole Sigma-Aldrich Imidocarb Dr. Ehrenstorfer Ipronidazole-OH -

The Grohe Method and Quinolone Antibiotics
The Grohe method and quinolone antibiotics Antibiotics are medicines that are used to treat bacterial for modern fluoroquinolones. The Grohe process and the infections. They contain active ingredients belonging to var- synthesis of ciprofloxacin sparked Bayer AG’s extensive ious substance classes, with modern fluoroquinolones one research on fluoroquinolones and the global competition of the most important and an indispensable part of both that produced additional potent antibiotics. human and veterinary medicine. It is largely thanks to Klaus Grohe – the “father of Bayer quinolones” – that this entirely In chemical terms, the antibiotics referred to for simplicity synthetic class of antibiotics now plays such a vital role for as quinolones are derived from 1,4-dihydro-4-oxo-3-quin- medical practitioners. From 1965 to 1997, Grohe worked oline carboxylic acid (1) substituted in position 1. as a chemist, carrying out basic research at Bayer AG’s Fluoroquinolones possess a fluorine atom in position 6. In main research laboratory (WHL) in Leverkusen. During this addition, ciprofloxacin (2) has a cyclopropyl group in posi- period, in 1975, he developed the Grohe process – a new tion 1 and also a piperazine group in position 7 (Figure A). multi-stage synthesis method for quinolones. It was this This substituent pattern plays a key role in its excellent achievement that first enabled him to synthesize active an- antibacterial efficacy. tibacterial substances such as ciprofloxacin – the prototype O 5 O 4 3 6 COOH F COOH 7 2 N N N 8 1 H N R (1) (2) Figure A: Basic structure of quinolone (1) (R = various substituents) and ciprofloxacin (2) Quinolones owe their antibacterial efficacy to their inhibition This unique mode of action also makes fluoroquinolones of essential bacterial enzymes – DNA gyrase (topoisomer- highly effective against a large number of pathogenic ase II) and topoisomerase IV. -
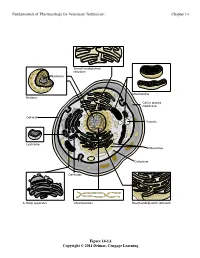
Fundamentals of Pharmacology for Veterinary Technicians Chapter 14
Fundamentals of Pharmacology for Veterinary Technicians Chapter 14 6PRRWKHQGRSODVPLF UHWLFXOXP 1XFOHROXV 0LWRFKRQGULD 1XFOHXV &HOORUSODVPD PHPEUDQH &HOOZDOO 9DFXROH /\VRVRPH 5LERVRPHV &\WRSODVP &HQWULROHV $*ROJLDSSDUDWXV &KURPRVRPHV 5RXJKHQGRSODVPLFUHWLFXOXP Figure 14-1A Copyright © 2011 Delmar, Cengage Learning Fundamentals of Pharmacology for Veterinary Technicians Chapter 14 1XFOHRLG '1$ 3LOXV 5LERVRPHV )ODJHOOXP &DSVXOH &HOOZDOO &HOOPHPEUDQH 3ODVPLG '1$ &\WRSODVP % Figure 14-1B Copyright © 2011 Delmar, Cengage Learning Fundamentals of Pharmacology for Veterinary Technicians Chapter 14 6WHS 3URFHGXUH 5HVXOW 3ULPDU\VWDLQ$SSO\FU\VWDO $OOEDFWHULDVWDLQSXUSOH YLROHWVWDLQ SXUSOH 5LQVHVOLGH 0RUGDQW$SSO\*UDP VLRGLQH $OOEDFWHULDUHPDLQSXUSOH 5LQVHVOLGH 'HFRORUL]H$SSO\DFLGDOFRKRORUDFHWRQH 3XUSOHVWDLQLVUHPRYHGIURP JUDPQHJDWLYHFHOOV 5LQVHVOLGH &RXQWHUVWDLQ$SSO\VDIUDQLQ *UDPQHJDWLYHFHOOVDSSHDU VWDLQ UHG SLQNUHGJUDPSRVLWLYHFHOOV DSSHDUSXUSOH 5LQVHVOLGH Figure 14-2 Copyright © 2011 Delmar, Cengage Learning Fundamentals of Pharmacology for Veterinary Technicians Chapter 14 Figure 14-3 Copyright © 2011 Delmar, Cengage Learning Fundamentals of Pharmacology for Veterinary Technicians Chapter 14 +JRIDQWLELRWLFSHUP/RIEURWK Figure 14-4 Copyright © 2011 Delmar, Cengage Learning Fundamentals of Pharmacology for Veterinary Technicians Chapter 14 1XFOHLFDFLG V\QWKHVLVDOWHUDWLRQ 1XFOHRLG '1$ 3URWHLQV\QWKHVLV LQKLELWLRQ 3LOXV 5LERVRPHV %ORFNLQJWKHXVH RIQHHGHGQXWULHQWV )ODJHOOXP &DSVXOH &HOOZDOO 3ODVPLG '1$ &\WRSODVP &HOOZDOO GLVUXSWLRQ &HOOPHPEUDQH &HOOPHPEUDQH -

Development and Validation of UV Spectrophotometric Method for Orbifloxacin Assay and Dissolution Studies
Brazilian Journal of Pharmaceutical Sciences vol. 50, n. 3, jul./sep., 2014 Article http://dx.doi.org/10.1590/S1984-82502014000300003 Development and validation of UV spectrophotometric method for orbifloxacin assay and dissolution studies Edith Cristina Laignier Cazedey, Hérida Regina Nunes Salgado* School of Pharmaceutical Sciences, State University of São Paulo “Júlio de Mesquita Filho”, Araraquara, São Paulo, Brazil New, simple and cost effective UV-spectrophotometric method was developed for the estimation of orbifloxacin in pharmaceutical formulation. Orbifloxacin was estimated at 290 nm in 0.5 M hydrochloric acid. Linearity range was found to be 1.0-6.0 μg mL–1. The method was tested and validated for various parameters according to main guidelines. The proposed method was successfully applied for the determination of orbifloxacin in tablets. The results demonstrated that the procedure is accurate, precise and reproducible, while being simple, economical and less time consuming. It can be suitably applied for the estimation of orbifloxacin in routine quality control and dissolution studies. Uniterms: Medicines/quality control. Medicines/dissolution. Orbifloxacin/determination. UV spectrophotometry/quantitative analysis/validation. Um método espectrofotométrico novo, simples e de baixo custo foi desenvolvido para a determinação de orbifloxacino em formulação farmacêutica. O orbifloxacino foi determinado em 290 nm utilizando ácido clorídrico 0,5 M como solvente. O intervalo de linearidade usado foi de 1,0 a 6,0 μg mL–1. O método foi testado e validado em vários parâmetros de acordo com os principais guias. O método proposto foi aplicado com sucesso para a determinação de orbifloxacino em comprimidos. Os resultados demonstraram que este procedimento é exato, preciso e reprodutível, ao mesmo tempo em que é simples, barato e de mais rápida execução e pode ser adequadamente aplicado para a determinação de orbifloxacino na rotina do controle de qualidade e em estudos de dissolução de comprimidos contendo este fármaco. -

Swedres-Svarm 2005
SVARM Swedish Veterinary Antimicrobial Resistance Monitoring Preface .............................................................................................4 Summary ..........................................................................................6 Sammanfattning................................................................................7 Use of antimicrobials (SVARM 2005) .................................................8 Resistance in zoonotic bacteria ......................................................13 Salmonella ..........................................................................................................13 Campylobacter ...................................................................................................16 Resistance in indicator bacteria ......................................................18 Escherichia coli ...................................................................................................18 Enterococcus .....................................................................................................22 Resistance in animal pathogens ......................................................32 Pig ......................................................................................................................32 Cattle ..................................................................................................................33 Horse ..................................................................................................................36 Dog -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -

Electronic Supplementary Material (ESI) for Analytical Methods. This Journal Is © the Royal Society of Chemistry 2014
Electronic Supplementary Material (ESI) for Analytical Methods. This journal is © The Royal Society of Chemistry 2014 Table 1 Mass spectrometer parameters and LOQ for 55 veterinary drugs EU MRLsa RTe Fragmentor LOQf(µg kg-1) NO Compound Group Transiton (CE) (µg kg-1) (min) (V) Porcine Bovine Ovine 1 Albendazole Benzimidazole 100c, 100d 17.1 266.1>234.1(20)*;266.1>191.1(35) 160 5 5 5 2 Albendazole sulfone Benzimidazole 100c, 100d 11.1 298.1>159.1(60)*;298.1>224.1(35) 140 2.4 2.4 2.4 3 Albendazole sulfoxide Benzimidazole 100c, 100d 8.1 282.1>240.1(15)*;282.1>208.1(40) 80 2.8 2.8 1.4 4 Albendazole-2-amino sulfone Benzimidazole 100c, 100d 4.4 240.1>133.1(35)*;240.1>198.1(10) 140 1 1 1 5 Febantel Benzimidazole 50b, 50c, 50d 26.2 447.2>383.1(10)*;447.2>415.1(15) 100 1.8 3.6 1.8 6 Flubendazole-amine Benzimidazole 50b 10.7 256.1>95.1(60)*;256.1>123.1(40) 160 2 2 2 7 Levamisole Benzimidazole 10c, 10d 4 205.1>178.1(20)*;205.1>91.1(45) 140 1 1 1 8 Mebendazole-5-hydroxy Benzimidazole 60d 9.8 298.1>266.1(15)*;298.1>79.1(40) 100 2 2 2 9 Mebendazole-amine Benzimidazole 60d 9.6 238.2>77.1(40)*;238.2>105.1(20) 160 2.4 1.2 1.2 10 Oxfendazole Benzimidazole 50b, 50c, 50d 12 316.1>159.1(25)*;316.1>191.1(10) 160 1 1 1 11 Oxibendazole Benzimidazole 100b 11.7 250.2>218.1(15)*;250.2>176.1(30) 80 10 10 10 12 Thiabendazole Benzimidazole 100c 5.78 202.1>175.0(30)*;202.1>131.1(35) 100 10 10 10 13 Thiabendazole-5-hydroxy Benzimidazole 100c, 100d 3.9 218.1>147.1(30)*;218.1>191.1(40) 120 1.7 1.7 1.7 14 Clindamycin Macrolide - 12.4 425.2>126.1(40)*;425.2>377.1(10) 120 -

Concept Paper
Concept Paper: Potential Approach for Ranking of Antimicrobial Drugs According to Their Importance in Human Medicine: A Risk Management Tool for Antimicrobial New Animal Drugs Introduction This concept paper discusses a potential approach to considering the human medical importance of antimicrobial drugs when assessing and managing antimicrobial resistance risks associated with the use of antimicrobial drugs in animals. In 2003, FDA established a list ranking antimicrobial drugs according to their relative importance in human medicine primarily for the purpose of supporting a qualitative risk assessment process outlined in agency guidance. It was envisioned by the Agency at the time the current medical importance rankings list was published that it would periodically reassess the rankings to align with contemporary science and current human clinical practices. To that end, this paper describes a potential revised process for ranking antimicrobial drugs according to their relative importance in human medicine, potential revised criteria to determine the medical importance rankings, and the list of antimicrobial drug medical importance rankings that would result if those criteria were to be used. Disclaimer: This concept paper is for discussion purposes only. The intent of this concept paper is to obtain public comment and early input on a potential approach to consider the human medical importance of antimicrobial drugs when assessing and managing antimicrobial resistance risks associated with the use of antimicrobial drugs in animals. This concept paper does not contain recommendations and does not constitute draft or final guidance by the Food and Drug Administration. It should not be used for any purpose other than to facilitate public comment. -

Belgian Veterinary Surveillance of Antimicrobial Consumption
Belgian Veterinary Surveillance of Antibacterial Consumption National consumption report 2018 Publication : 19 June 2019 1 SUMMARY This annual BelVet-SAC report is now published for the 10th time and describes the antibacterial use in animals in Belgium in 2018 and the evolution since 2011. For the first time this report combines sales data (collected at the level of the wholesalers- distributors and the compound feed producers) and usage data (collected at herd level). This allows to dig deeper into AMU at species and herd level in Belgium. With -12,8% mg antimicrobial/kg biomass in comparison to 2017, 2018 marks the largest reduction in total sales of antimicrobials for animals in Belgium since 2011. This obviously continues the decreasing trend of the previous years, resulting in a cumulative reduction of -35,4% mg/kg since 2011. This reduction is evenly split over a reduction in pharmaceuticals (-13,2% mg/kg) and antibacterial premixes (-9,2% mg/kg). It is speculated that the large reduction observed in 2018 might partly be due to the effect of extra stock (of pharmaceuticals) taken during 2017 by wholesalers-distributors and veterinarians in anticipation of the increase in the antimicrobial tax for Marketing Authorisation Holders, which became effective on the 1st of April 2018. When comparing the results achieved in 2018 with the AMCRA 2020 reduction targets, the goal of reducing the overall AMU in animals with 50% by 2020 has not been achieved yet, however, the objective comes in range with still 14,6% to reduce over the next two years. Considering the large reduction observed in total AMU in 2018, it is not surprising that also in the pig sector a substantial reduction of -8,3% mg/kg between 2017 and 2018 is observed based upon the usage data.