Python in Computational Neuroscience & Modular Toolkit for Data Processing (MDP)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Practice Tips for Open Source Licensing Adam Kubelka
Santa Clara High Technology Law Journal Volume 22 | Issue 4 Article 4 2006 No Free Beer - Practice Tips for Open Source Licensing Adam Kubelka Matthew aF wcett Follow this and additional works at: http://digitalcommons.law.scu.edu/chtlj Part of the Law Commons Recommended Citation Adam Kubelka and Matthew Fawcett, No Free Beer - Practice Tips for Open Source Licensing, 22 Santa Clara High Tech. L.J. 797 (2005). Available at: http://digitalcommons.law.scu.edu/chtlj/vol22/iss4/4 This Article is brought to you for free and open access by the Journals at Santa Clara Law Digital Commons. It has been accepted for inclusion in Santa Clara High Technology Law Journal by an authorized administrator of Santa Clara Law Digital Commons. For more information, please contact [email protected]. ARTICLE NO FREE BEER - PRACTICE TIPS FOR OPEN SOURCE LICENSING Adam Kubelkat Matthew Fawcetttt I. INTRODUCTION Open source software is big business. According to research conducted by Optaros, Inc., and InformationWeek magazine, 87 percent of the 512 companies surveyed use open source software, with companies earning over $1 billion in annual revenue saving an average of $3.3 million by using open source software in 2004.1 Open source is not just staying in computer rooms either-it is increasingly grabbing intellectual property headlines and entering mainstream news on issues like the following: i. A $5 billion dollar legal dispute between SCO Group Inc. (SCO) and International Business Machines Corp. t Adam Kubelka is Corporate Counsel at JDS Uniphase Corporation, where he advises the company on matters related to the commercialization of its products. -

ACS – the Archival Cytometry Standard
http://flowcyt.sf.net/acs/latest.pdf ACS – the Archival Cytometry Standard Archival Cytometry Standard ACS International Society for Advancement of Cytometry Candidate Recommendation DRAFT Document Status The Archival Cytometry Standard (ACS) has undergone several revisions since its initial development in June 2007. The current proposal is an ISAC Candidate Recommendation Draft. It is assumed, however not guaranteed, that significant features and design aspects will remain unchanged for the final version of the Recommendation. This specification has been formally tested to comply with the W3C XML schema version 1.0 specification but no position is taken with respect to whether a particular software implementing this specification performs according to medical or other valid regulations. The work may be used under the terms of the Creative Commons Attribution-ShareAlike 3.0 Unported license. You are free to share (copy, distribute and transmit), and adapt the work under the conditions specified at http://creativecommons.org/licenses/by-sa/3.0/legalcode. Disclaimer of Liability The International Society for Advancement of Cytometry (ISAC) disclaims liability for any injury, harm, or other damage of any nature whatsoever, to persons or property, whether direct, indirect, consequential or compensatory, directly or indirectly resulting from publication, use of, or reliance on this Specification, and users of this Specification, as a condition of use, forever release ISAC from such liability and waive all claims against ISAC that may in any manner arise out of such liability. ISAC further disclaims all warranties, whether express, implied or statutory, and makes no assurances as to the accuracy or completeness of any information published in the Specification. -

Press Release: New and Revised Extensions for Accessible
Press release Leuven, Belgium, 8 November 2011 New and Revised Extensions for Accessible Document Creation with OpenOffice.org and LibreOffice The Katholieke Universiteit Leuven (K.U.Leuven) today released an extension for OpenOffice.org Writer and LibreOffice Writer that enables users to evaluate and repair accessibility issues in word processing documents. “AccessODF” (http://sourceforge.net/p/accessodf/wiki/) is a freeware extension for OpenOffice.org and LibreOffice, two office suites that are freely available for Microsoft Windows, Mac OS X, Linux/Unix and Solaris. At the same time, K.U.Leuven also releases new versions of two other extensions: odt2daisy (http://odt2daisy.sourceforge.net/) and odt2braille (http://odt2braille.sourceforge.net/). The former enables users to export word processing documents to digital talking books in the DAISY format; the latter enables exporting to Braille and printing on a Braille embosser. AccessODF, odt2daisy and odt2braille are being developed in the framework of the AEGIS project, an R&D project funded by the European Commission. The three extensions will be demonstrated at the AEGIS project’s Workshop and Conference, which take place in Brussels on 28-30 November 2011 (http://aegis-conference.eu/). AccessODF AccessODF is an extension that can be used in OpenOffice.org Writer and in LibreOffice Writer. It enables authors to find and repair accessibility issues in their documents, i.e. issues that make their documents difficult or even impossible to read for people with disabilities. This includes -

Pack, Encrypt, Authenticate Document Revision: 2021 05 02
PEA Pack, Encrypt, Authenticate Document revision: 2021 05 02 Author: Giorgio Tani Translation: Giorgio Tani This document refers to: PEA file format specification version 1 revision 3 (1.3); PEA file format specification version 2.0; PEA 1.01 executable implementation; Present documentation is released under GNU GFDL License. PEA executable implementation is released under GNU LGPL License; please note that all units provided by the Author are released under LGPL, while Wolfgang Ehrhardt’s crypto library units used in PEA are released under zlib/libpng License. PEA file format and PCOMPRESS specifications are hereby released under PUBLIC DOMAIN: the Author neither has, nor is aware of, any patents or pending patents relevant to this technology and do not intend to apply for any patents covering it. As far as the Author knows, PEA file format in all of it’s parts is free and unencumbered for all uses. Pea is on PeaZip project official site: https://peazip.github.io , https://peazip.org , and https://peazip.sourceforge.io For more information about the licenses: GNU GFDL License, see http://www.gnu.org/licenses/fdl.txt GNU LGPL License, see http://www.gnu.org/licenses/lgpl.txt 1 Content: Section 1: PEA file format ..3 Description ..3 PEA 1.3 file format details ..5 Differences between 1.3 and older revisions ..5 PEA 2.0 file format details ..7 PEA file format’s and implementation’s limitations ..8 PCOMPRESS compression scheme ..9 Algorithms used in PEA format ..9 PEA security model .10 Cryptanalysis of PEA format .12 Data recovery from -
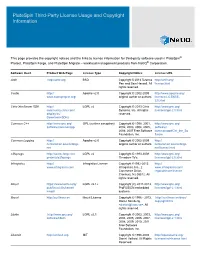
Platespin Third-Party License Usage and Copyright Information
PlateSpin Third-Party License Usage and Copyright Information This page provides the copyright notices and the links to license information for third-party software used in PlateSpin® Protect, PlateSpin Forge, and PlateSpin Migrate – workload management products from NetIQ® Corporation. Software Used Product Web Page License Type Copyright Notice License URL Antlr http://antlr.org BSD Copyright © 2012 Terence http://antlr.org/ Parr and Sam Harwell. All license.html rights reserved. Castle http:// Apache v2.0 Copyright © 2002-2009 http://www.apache.org/ www.castleproject.org/ original author or authors. licenses/LICENSE- 2.0.html Citrix XenServer SDK http:// LGPL v2 Copyright © 2013 Citrix http://www.gnu.org/ community.citrix.com/ Systems, Inc. All rights licenses/lgpl-2.0.html display/xs/ reserved. Download+SDKs Common C++ http://www.gnu.org/ GPL (runtime exception) Copyright © 1998, 2001, http://www.gnu.org/ software/commoncpp 2002, 2003, 2004, 2005, software/ 2006, 2007 Free Software commoncpp#Get_the_So Foundation, Inc. ftware Common.Logging http:// Apache v2.0 Copyright © 2002-2009 http:// netcommon.sourceforge. original author or authors. netcommon.sourceforge. net net/license.html e2fsprogs http://sourceforge.net/ LGPL v2 Copyright © 1993-2008 http://www.gnu.org/ projects/e2fsprogs Theodore Ts'o. licenses/lgpl-2.0.html Infragistics http:// Infragistics License Copyright ©1992-2013 http:// www.infragistics.com Infragistics, Inc., 2 www.infragistics.com/ Commerce Drive, legal/ultimate/license Cranbury, NJ 08512. All rights reserved. Kmod https://www.kernel.org/ LGPL v2.1+ Copyright (C) 2011-2013 http://www.gnu.org/ pub/linux/utils/kernel/ ProFUSION embedded licenses/lgpl-2.1.html kmod/ systems libcurl http://curl.haxx.se libcurl License Copyright © 1996 - 2013, http://curl.haxx.se/docs/ Daniel Stenberg, copyright.html <[email protected]>. -

Are Projects Hosted on Sourceforge.Net Representative of the Population of Free/Open Source Software Projects?
Are Projects Hosted on Sourceforge.net Representative of the Population of Free/Open Source Software Projects? Bob English February 5, 2008 Introduction We are researchers studying Free/Open Source Software (FOSS) projects . Because some of our work uses data gathered from Sourceforge.net (SF), a web site that hosts FOSS projects, we consider it important to understand how representative SF is of the entire population of FOSS projects. While SF is probably hosts the greatest number of FOSS projects, there are many other similar hosting sites. In addition, many FOSS projects maintain their own web sites and other project infrastructure on their own servers. Based on performing an Internet search and literature review (see Appendix for search procedures), it appears that no one knows how many Free/Open Source Software Projects exist in the world or how many people are working on them. Because the population of FOSS projects and developers is unknown, it is not surprising that we were not able to find any empirical analysis to assess how representative SF is of this unknown population. Although an extensive empirical analysis has not been performed, many researchers have considered the question or related questions. The next section of this paper reviews some of the FOSS literature that discusses the representativeness of SF. The third section of the paper describes some ideas for empirically exploring the representativeness of SF. The paper closes with some arguments for believing SF is the best choice for those interested in studying the population of FOSS projects. Literature Review In one of the earliest references to the representativeness of SF, Madley, Freeh and Tynan (2002) mention that they assumed that SF was representative because of its popularity and because of the number of projects hosted there, although they note that this assumption “needs to be confirmed” (p. -

Reactos-Devtutorial.Pdf
Developer Tutorials Developer Tutorials Next Developer Tutorials Table of Contents I. Newbie Developer 1. Introduction to ReactOS development 2. Where to get the latest ReactOS source, compilation tools and how to compile the source 3. Testing your compiled ReactOS code 4. Where to go from here (newbie developer) II. Centralized Source Code Repository 5. Introducing CVS 6. Downloading and configuring your CVS client 7. Checking out a new tree 8. Updating your tree with the latest code 9. Applying for write access 10. Submitting your code with CVS 11. Submitting a patch to the project III. Advanced Developer 12. CD Packaging Guide 13. ReactOS Architecture Whitepaper 14. ReactOS WINE Developer Guide IV. Bochs testing 15. Introducing Bochs 16. Downloading and Using Bochs with ReactOS 17. The compile, test and debug cycle under Bochs V. VMware Testing 18. Introducing VMware List of Tables 7.1. Modules http://reactos.com/rosdocs/tutorials/bk02.html (1 of 2) [3/18/2003 12:16:53 PM] Developer Tutorials Prev Up Next Chapter 8. Where to go from here Home Part I. Newbie Developer (newbie user) http://reactos.com/rosdocs/tutorials/bk02.html (2 of 2) [3/18/2003 12:16:53 PM] Part I. Newbie Developer Part I. Newbie Developer Prev Developer Tutorials Next Newbie Developer Table of Contents 1. Introduction to ReactOS development 2. Where to get the latest ReactOS source, compilation tools and how to compile the source 3. Testing your compiled ReactOS code 4. Where to go from here (newbie developer) Prev Up Next Developer Tutorials Home Chapter 1. Introduction to ReactOS development http://reactos.com/rosdocs/tutorials/bk02pt01.html [3/18/2003 12:16:54 PM] Chapter 1. -

The Best Things in Life Are Free by Jeff Martin
Administration | Column The Best Things in Life are Free by Jeff Martin he best things in life are free. Love, respect, software developers.) Every water utility should be Tfriendship, family, forgiveness, down-home aware of the savings opportunity, tight budget or not. goodness, and grandma’s apple pie cannot be The challenge is that offers of free software can include purchased. Even the Beatles sang “Can’t Buy Me Love.” great value, but other times often include great danger These blessings can only be given. and a hefty price. Yet what about all the ‘free’ offers today? Can they be Many offers of free software are truly a dangerous trusted? Free vacations (with a 90-minute time-share hook covered with bait. The free or low cost software presentation)? Free samples at the grocery store? Free promises a solution, but in the end the promise magazine subscriptions? Free websites like www. conceals a hidden agenda. At the worst, ‘malware’ is freecycle.org and www.craigslist.org? Free classified software that secretly contains a virus that is designed ads? Free shipping? Free money? Free education? to harm, steal information, or even take your computer Ultimately nothing is free for someone somewhere is hostage for a ransom. There is also ‘adware’ which paying the bottom line. I only remember one thing may provide the promised software function, but also from high school economics, “TANSTAAFL” was requires the viewing of advertisements. Some ‘adware’ posted above the door. “There ain’t no such thing as a is actually respectable and so this is not necessarily a free lunch.” Beware the hook beneath the bait! show stopper. -

Jsignpdf Quick Start Guide
JSignPdf Quick Start Guide Josef Cacek 2.0.0 Table of Contents JSignPdf Introduction . 1 Benefits of digital signatures. 1 License . 1 History . 2 Author . 2 Getting support . 2 Prerequisites . 3 Java . 3 Keystore . 3 Launching. 4 Using JSignPdf – signing PDF files. 5 Simple version . 5 More detailed version . 5 Advanced view . 6 Encryption . 8 Visible signature. 9 TSA – timestamps . 11 Certificate revocation checking . 11 Proxy settings . 12 Using hardware tokens for signing . 13 Advanced application configuration . 14 conf.properties . 14 Java VM options using EXE launchers . 14 Solving problems . 15 Out of memory error. 15 Command line (batch mode) . 16 Program exit codes . 19 Examples . 20 Other command line tools . 21 InstallCert Tool . 21 JSignPdf Introduction JSignPdf is an open-source application that adds digital signatures to PDF documents. It’s written in Java programming language and it can be launched on the most of current OS. Users can control the application using simple GUI or command line arguments. Main features: • supports visible signatures • can set certification level • supports PDF encryption with setting rights • timestamp support • certificate revocation checking (CRL and/or OCSP) Benefits of digital signatures Below are some common reasons for applying a digital signature to communications. (source Wikipedia) Authentication Although messages may often include information about the entity sending a message, that information may not be accurate. Digital signatures can be used to authenticate the source of messages. When ownership of a digital signature secret key is bound to a specific user, a valid signature shows that the message was sent by that user. The importance of high confidence in sender authenticity is especially obvious in a financial context. -

The Open Source Software Development Phenomenon: an Analysis Based on Social Network Theory
THE OPEN SOURCE SOFTWARE DEVELOPMENT PHENOMENON: AN ANALYSIS BASED ON SOCIAL NETWORK THEORY Greg Madey Vincent Freeh Computer Science & Engineering Computer Science & Engineering University of Notre Dame University of Notre Dame [email protected] [email protected] Renee Tynan Department of Management University of Notre Dame [email protected] Abstract The OSS movement is a phenomenon that challenges many traditional theories in economics, software engineering, business strategy, and IT management. Thousands of software programmers are spending tremendous amounts of time and effort writing and debugging software, most often with no direct monetary compensation. The programs, some of which are extremely large and complex, are written without the benefit of traditional project management, change tracking, or error checking techniques. Since the programmers are working outside of a traditional organizational reward structure, accountability is an issue as well. A significant portion of internet e-commerce runs on OSS, and thus many firms have little choice but to trust mission-critical e-commerce systems to run on such software, requiring IT management to deal with new types of socio-technical problems. A better understanding of how the OSS community functions may help IT planners make more informed decisions and develop more effective strategies for using OSS software. We hypothesize that open source software development can be modeled as self-organizing, collaboration, social networks. We analyze structural data on over 39,000 open source projects hosted at SourceForge.net involving over 33,000 developers. We define two software developers to be connected — part of a collaboration social network — if they are members of the same project, or are connected by a chain of connected developers. -

Software Requirements Specification
Software Requirements Specification for PeaZip Requirements for version 2.7.1 Prepared by Liles Athanasios-Alexandros Software Engineering , AUTH 12/19/2009 Software Requirements Specification for PeaZip Page ii Table of Contents Table of Contents.......................................................................................................... ii 1. Introduction.............................................................................................................. 1 1.1 Purpose ........................................................................................................................1 1.2 Document Conventions.................................................................................................1 1.3 Intended Audience and Reading Suggestions...............................................................1 1.4 Project Scope ...............................................................................................................1 1.5 References ...................................................................................................................2 2. Overall Description .................................................................................................. 3 2.1 Product Perspective......................................................................................................3 2.2 Product Features ..........................................................................................................4 2.3 User Classes and Characteristics .................................................................................5 -

Libreoffice Base Guide 6.4 | 3 Relationships Between Tables
Copyright This document is Copyright © 2020 by the LibreOffice Documentation Team. Contributors are listed below. You may distribute it and/or modify it under the terms of either the GNU General Public License (http://www.gnu.org/licenses/gpl.html), version 3 or later, or the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), version 4.0 or later. All trademarks within this guide belong to their legitimate owners. Contributors This guide has been updated from Base Guide 6.2. To this edition Pulkit Krishna Dan Lewis Jenna Sargent Drew Jensen Jean-Pierre Ledure Jean Hollis Weber To previous editions Pulkit Krishna Jean Hollis Weber Dan Lewis Peter Scholfield Jochen Schiffers Robert Großkopf Jost Lange Martin Fox Hazel Russman Steve Schwettman Alain Romedenne Andrew Pitonyak Jean-Pierre Ledure Drew Jensen Randolph GAMO Feedback Please direct any comments or suggestions about this document to the Documentation Team’s mailing list: [email protected] Note Everything you send to a mailing list, including your email address and any other personal information that is written in the message, is publicly archived and cannot be deleted. Publication date and software version Published July 2020. Based on LibreOffice 6.4. Documentation for LibreOffice is available at http://documentation.libreoffice.org/en/ Contents Copyright.....................................................................................................................................2 Contributors............................................................................................................................2