Pymol User's Guide
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Practice Tips for Open Source Licensing Adam Kubelka
Santa Clara High Technology Law Journal Volume 22 | Issue 4 Article 4 2006 No Free Beer - Practice Tips for Open Source Licensing Adam Kubelka Matthew aF wcett Follow this and additional works at: http://digitalcommons.law.scu.edu/chtlj Part of the Law Commons Recommended Citation Adam Kubelka and Matthew Fawcett, No Free Beer - Practice Tips for Open Source Licensing, 22 Santa Clara High Tech. L.J. 797 (2005). Available at: http://digitalcommons.law.scu.edu/chtlj/vol22/iss4/4 This Article is brought to you for free and open access by the Journals at Santa Clara Law Digital Commons. It has been accepted for inclusion in Santa Clara High Technology Law Journal by an authorized administrator of Santa Clara Law Digital Commons. For more information, please contact [email protected]. ARTICLE NO FREE BEER - PRACTICE TIPS FOR OPEN SOURCE LICENSING Adam Kubelkat Matthew Fawcetttt I. INTRODUCTION Open source software is big business. According to research conducted by Optaros, Inc., and InformationWeek magazine, 87 percent of the 512 companies surveyed use open source software, with companies earning over $1 billion in annual revenue saving an average of $3.3 million by using open source software in 2004.1 Open source is not just staying in computer rooms either-it is increasingly grabbing intellectual property headlines and entering mainstream news on issues like the following: i. A $5 billion dollar legal dispute between SCO Group Inc. (SCO) and International Business Machines Corp. t Adam Kubelka is Corporate Counsel at JDS Uniphase Corporation, where he advises the company on matters related to the commercialization of its products. -

Mercury 2.4 User Guide and Tutorials 2011 CSDS Release
Mercury 2.4 User Guide and Tutorials 2011 CSDS Release Copyright © 2010 The Cambridge Crystallographic Data Centre Registered Charity No 800579 Conditions of Use The Cambridge Structural Database System (CSD System) comprising all or some of the following: ConQuest, Quest, PreQuest, Mercury, (Mercury CSD and Materials module of Mercury), VISTA, Mogul, IsoStar, SuperStar, web accessible CSD tools and services, WebCSD, CSD Java sketcher, CSD data file, CSD-UNITY, CSD-MDL, CSD-SDfile, CSD data updates, sub files derived from the foregoing data files, documentation and command procedures (each individually a Component) is a database and copyright work belonging to the Cambridge Crystallographic Data Centre (CCDC) and its licensors and all rights are protected. Use of the CSD System is permitted solely in accordance with a valid Licence of Access Agreement and all Components included are proprietary. When a Component is supplied independently of the CSD System its use is subject to the conditions of the separate licence. All persons accessing the CSD System or its Components should make themselves aware of the conditions contained in the Licence of Access Agreement or the relevant licence. In particular: • The CSD System and its Components are licensed subject to a time limit for use by a specified organisation at a specified location. • The CSD System and its Components are to be treated as confidential and may NOT be disclosed or re- distributed in any form, in whole or in part, to any third party. • Software or data derived from or developed using the CSD System may not be distributed without prior written approval of the CCDC. -

Open Babel Documentation Release 2.3.1
Open Babel Documentation Release 2.3.1 Geoffrey R Hutchison Chris Morley Craig James Chris Swain Hans De Winter Tim Vandermeersch Noel M O’Boyle (Ed.) December 05, 2011 Contents 1 Introduction 3 1.1 Goals of the Open Babel project ..................................... 3 1.2 Frequently Asked Questions ....................................... 4 1.3 Thanks .................................................. 7 2 Install Open Babel 9 2.1 Install a binary package ......................................... 9 2.2 Compiling Open Babel .......................................... 9 3 obabel and babel - Convert, Filter and Manipulate Chemical Data 17 3.1 Synopsis ................................................. 17 3.2 Options .................................................. 17 3.3 Examples ................................................. 19 3.4 Differences between babel and obabel .................................. 21 3.5 Format Options .............................................. 22 3.6 Append property values to the title .................................... 22 3.7 Filtering molecules from a multimolecule file .............................. 22 3.8 Substructure and similarity searching .................................. 25 3.9 Sorting molecules ............................................ 25 3.10 Remove duplicate molecules ....................................... 25 3.11 Aliases for chemical groups ....................................... 26 4 The Open Babel GUI 29 4.1 Basic operation .............................................. 29 4.2 Options ................................................. -

ACS – the Archival Cytometry Standard
http://flowcyt.sf.net/acs/latest.pdf ACS – the Archival Cytometry Standard Archival Cytometry Standard ACS International Society for Advancement of Cytometry Candidate Recommendation DRAFT Document Status The Archival Cytometry Standard (ACS) has undergone several revisions since its initial development in June 2007. The current proposal is an ISAC Candidate Recommendation Draft. It is assumed, however not guaranteed, that significant features and design aspects will remain unchanged for the final version of the Recommendation. This specification has been formally tested to comply with the W3C XML schema version 1.0 specification but no position is taken with respect to whether a particular software implementing this specification performs according to medical or other valid regulations. The work may be used under the terms of the Creative Commons Attribution-ShareAlike 3.0 Unported license. You are free to share (copy, distribute and transmit), and adapt the work under the conditions specified at http://creativecommons.org/licenses/by-sa/3.0/legalcode. Disclaimer of Liability The International Society for Advancement of Cytometry (ISAC) disclaims liability for any injury, harm, or other damage of any nature whatsoever, to persons or property, whether direct, indirect, consequential or compensatory, directly or indirectly resulting from publication, use of, or reliance on this Specification, and users of this Specification, as a condition of use, forever release ISAC from such liability and waive all claims against ISAC that may in any manner arise out of such liability. ISAC further disclaims all warranties, whether express, implied or statutory, and makes no assurances as to the accuracy or completeness of any information published in the Specification. -

Press Release: New and Revised Extensions for Accessible
Press release Leuven, Belgium, 8 November 2011 New and Revised Extensions for Accessible Document Creation with OpenOffice.org and LibreOffice The Katholieke Universiteit Leuven (K.U.Leuven) today released an extension for OpenOffice.org Writer and LibreOffice Writer that enables users to evaluate and repair accessibility issues in word processing documents. “AccessODF” (http://sourceforge.net/p/accessodf/wiki/) is a freeware extension for OpenOffice.org and LibreOffice, two office suites that are freely available for Microsoft Windows, Mac OS X, Linux/Unix and Solaris. At the same time, K.U.Leuven also releases new versions of two other extensions: odt2daisy (http://odt2daisy.sourceforge.net/) and odt2braille (http://odt2braille.sourceforge.net/). The former enables users to export word processing documents to digital talking books in the DAISY format; the latter enables exporting to Braille and printing on a Braille embosser. AccessODF, odt2daisy and odt2braille are being developed in the framework of the AEGIS project, an R&D project funded by the European Commission. The three extensions will be demonstrated at the AEGIS project’s Workshop and Conference, which take place in Brussels on 28-30 November 2011 (http://aegis-conference.eu/). AccessODF AccessODF is an extension that can be used in OpenOffice.org Writer and in LibreOffice Writer. It enables authors to find and repair accessibility issues in their documents, i.e. issues that make their documents difficult or even impossible to read for people with disabilities. This includes -

Real-Time Pymol Visualization for Rosetta and Pyrosetta
Real-Time PyMOL Visualization for Rosetta and PyRosetta Evan H. Baugh1, Sergey Lyskov1, Brian D. Weitzner1, Jeffrey J. Gray1,2* 1 Department of Chemical and Biomolecular Engineering, The Johns Hopkins University, Baltimore, Maryland, United States of America, 2 Program in Molecular Biophysics, The Johns Hopkins University, Baltimore, Maryland, United States of America Abstract Computational structure prediction and design of proteins and protein-protein complexes have long been inaccessible to those not directly involved in the field. A key missing component has been the ability to visualize the progress of calculations to better understand them. Rosetta is one simulation suite that would benefit from a robust real-time visualization solution. Several tools exist for the sole purpose of visualizing biomolecules; one of the most popular tools, PyMOL (Schro¨dinger), is a powerful, highly extensible, user friendly, and attractive package. Integrating Rosetta and PyMOL directly has many technical and logistical obstacles inhibiting usage. To circumvent these issues, we developed a novel solution based on transmitting biomolecular structure and energy information via UDP sockets. Rosetta and PyMOL run as separate processes, thereby avoiding many technical obstacles while visualizing information on-demand in real-time. When Rosetta detects changes in the structure of a protein, new coordinates are sent over a UDP network socket to a PyMOL instance running a UDP socket listener. PyMOL then interprets and displays the molecule. This implementation also allows remote execution of Rosetta. When combined with PyRosetta, this visualization solution provides an interactive environment for protein structure prediction and design. Citation: Baugh EH, Lyskov S, Weitzner BD, Gray JJ (2011) Real-Time PyMOL Visualization for Rosetta and PyRosetta. -

Instructions on Making Pdf Files Containing 3D
Detailed Instructions for Creating an Embedded Virtual 3-D Image of a Molecule Creating the documents with embedded virtual three-dimensional images may be done fairly easily by repeating the following procedures. Once you have embedded your first image, the use of that image is limited only by your imagination. The following instructions are written in extreme detail with annotated screen shots of the four programs you will use throughout for clarification. Once you are familiar with the procedure embedding a structure of interest should take less than 20 min., limited only by the speed with which you can click a mouse. In practice most of our embedded 3-D images have been created by our undergraduate student collaborators. Step 1. Creating a 2-D image in ChemSketch Using ACD ChemSketch 11.0 Freeware (http://www.acdlabs.com/download/chemsk_download.html ) a two dimensional structure may be drawn as per your specific need (see Figure 1). Once the molecule is drawn, optimized it by clicking the 3-D optimization button then save as an MDL molfile (*.mol). Figure 1: A screen shot of acetone drawn with ChemSketch after 3-D optimization. Step 2. Converting the *.mol file to a *.pdb file using Open Babel The file can now be converted from a .mol file to a .pdb file, Protein Data Bank format, to be properly accessed with the Adobe Acrobat 3D Toolkit 8.1.0. The file conversion is done using Open Babel Graphical User Interface v2.2.0 (openbabel.sourceforge.net) under default settings. Step by step instructions: A. -

Getting Started in Jmol
Getting Started in Jmol Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingResources/jmol/jmolTraining/started.html Introduction Physical models of proteins are powerful tools that can be used synergistically with computer visualizations to explore protein structure and function. Although it is interesting to explore models and visualizations created by others, it is much more engaging to create your own! At the MSOE Center for BioMolecular Modeling we use the molecular visualization program Jmol to explore protein and molecular structures in fully interactive 3-dimensional displays. Jmol a free, open source molecular visualization program used by students, educators and researchers internationally. The Jmol Training Guide from the MSOE Center for BioMolecular Modeling will provide the tools needed to create molecular renderings, physical models using 3-D printing technologies, as well as Jmol animations for online tutorials or electronic posters. Examples of Proteins in Jmol Jmol allows users to rotate proteins and molecular structures in a fully interactive 3-dimensional display. Some sample proteins designed with Jmol are shown to the right. Hemoglobin Proteins Insulin Proteins Green Fluorescent safely carry oxygen in the help regulate sugar in Proteins create blood. the bloodstream. bioluminescence in animals like jellyfish. Downloading Jmol Jmol Can be Used in Two Ways: 1. As an independent program on a desktop - Jmol can be downloaded to run on your desktop like any other program. It uses a Java platform and therefore functions equally well in a PC or Mac environment. 2. As a web application - Jmol has a web-based version (oftern refered to as "JSmol") that runs on a JavaScript platform and therefore functions equally well on all HTML5 compatible browsers such as Firefox, Internet Explorer, Safari and Chrome. -

Pack, Encrypt, Authenticate Document Revision: 2021 05 02
PEA Pack, Encrypt, Authenticate Document revision: 2021 05 02 Author: Giorgio Tani Translation: Giorgio Tani This document refers to: PEA file format specification version 1 revision 3 (1.3); PEA file format specification version 2.0; PEA 1.01 executable implementation; Present documentation is released under GNU GFDL License. PEA executable implementation is released under GNU LGPL License; please note that all units provided by the Author are released under LGPL, while Wolfgang Ehrhardt’s crypto library units used in PEA are released under zlib/libpng License. PEA file format and PCOMPRESS specifications are hereby released under PUBLIC DOMAIN: the Author neither has, nor is aware of, any patents or pending patents relevant to this technology and do not intend to apply for any patents covering it. As far as the Author knows, PEA file format in all of it’s parts is free and unencumbered for all uses. Pea is on PeaZip project official site: https://peazip.github.io , https://peazip.org , and https://peazip.sourceforge.io For more information about the licenses: GNU GFDL License, see http://www.gnu.org/licenses/fdl.txt GNU LGPL License, see http://www.gnu.org/licenses/lgpl.txt 1 Content: Section 1: PEA file format ..3 Description ..3 PEA 1.3 file format details ..5 Differences between 1.3 and older revisions ..5 PEA 2.0 file format details ..7 PEA file format’s and implementation’s limitations ..8 PCOMPRESS compression scheme ..9 Algorithms used in PEA format ..9 PEA security model .10 Cryptanalysis of PEA format .12 Data recovery from -
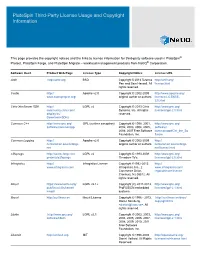
Platespin Third-Party License Usage and Copyright Information
PlateSpin Third-Party License Usage and Copyright Information This page provides the copyright notices and the links to license information for third-party software used in PlateSpin® Protect, PlateSpin Forge, and PlateSpin Migrate – workload management products from NetIQ® Corporation. Software Used Product Web Page License Type Copyright Notice License URL Antlr http://antlr.org BSD Copyright © 2012 Terence http://antlr.org/ Parr and Sam Harwell. All license.html rights reserved. Castle http:// Apache v2.0 Copyright © 2002-2009 http://www.apache.org/ www.castleproject.org/ original author or authors. licenses/LICENSE- 2.0.html Citrix XenServer SDK http:// LGPL v2 Copyright © 2013 Citrix http://www.gnu.org/ community.citrix.com/ Systems, Inc. All rights licenses/lgpl-2.0.html display/xs/ reserved. Download+SDKs Common C++ http://www.gnu.org/ GPL (runtime exception) Copyright © 1998, 2001, http://www.gnu.org/ software/commoncpp 2002, 2003, 2004, 2005, software/ 2006, 2007 Free Software commoncpp#Get_the_So Foundation, Inc. ftware Common.Logging http:// Apache v2.0 Copyright © 2002-2009 http:// netcommon.sourceforge. original author or authors. netcommon.sourceforge. net net/license.html e2fsprogs http://sourceforge.net/ LGPL v2 Copyright © 1993-2008 http://www.gnu.org/ projects/e2fsprogs Theodore Ts'o. licenses/lgpl-2.0.html Infragistics http:// Infragistics License Copyright ©1992-2013 http:// www.infragistics.com Infragistics, Inc., 2 www.infragistics.com/ Commerce Drive, legal/ultimate/license Cranbury, NJ 08512. All rights reserved. Kmod https://www.kernel.org/ LGPL v2.1+ Copyright (C) 2011-2013 http://www.gnu.org/ pub/linux/utils/kernel/ ProFUSION embedded licenses/lgpl-2.1.html kmod/ systems libcurl http://curl.haxx.se libcurl License Copyright © 1996 - 2013, http://curl.haxx.se/docs/ Daniel Stenberg, copyright.html <[email protected]>. -

Are Projects Hosted on Sourceforge.Net Representative of the Population of Free/Open Source Software Projects?
Are Projects Hosted on Sourceforge.net Representative of the Population of Free/Open Source Software Projects? Bob English February 5, 2008 Introduction We are researchers studying Free/Open Source Software (FOSS) projects . Because some of our work uses data gathered from Sourceforge.net (SF), a web site that hosts FOSS projects, we consider it important to understand how representative SF is of the entire population of FOSS projects. While SF is probably hosts the greatest number of FOSS projects, there are many other similar hosting sites. In addition, many FOSS projects maintain their own web sites and other project infrastructure on their own servers. Based on performing an Internet search and literature review (see Appendix for search procedures), it appears that no one knows how many Free/Open Source Software Projects exist in the world or how many people are working on them. Because the population of FOSS projects and developers is unknown, it is not surprising that we were not able to find any empirical analysis to assess how representative SF is of this unknown population. Although an extensive empirical analysis has not been performed, many researchers have considered the question or related questions. The next section of this paper reviews some of the FOSS literature that discusses the representativeness of SF. The third section of the paper describes some ideas for empirically exploring the representativeness of SF. The paper closes with some arguments for believing SF is the best choice for those interested in studying the population of FOSS projects. Literature Review In one of the earliest references to the representativeness of SF, Madley, Freeh and Tynan (2002) mention that they assumed that SF was representative because of its popularity and because of the number of projects hosted there, although they note that this assumption “needs to be confirmed” (p. -

Chemdoodle Web Components: HTML5 Toolkit for Chemical Graphics, Interfaces, and Informatics Melanie C Burger1,2*
Burger. J Cheminform (2015) 7:35 DOI 10.1186/s13321-015-0085-3 REVIEW Open Access ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics Melanie C Burger1,2* Abstract ChemDoodle Web Components (abbreviated CWC, iChemLabs, LLC) is a light-weight (~340 KB) JavaScript/HTML5 toolkit for chemical graphics, structure editing, interfaces, and informatics based on the proprietary ChemDoodle desktop software. The library uses <canvas> and WebGL technologies and other HTML5 features to provide solutions for creating chemistry-related applications for the web on desktop and mobile platforms. CWC can serve a broad range of scientific disciplines including crystallography, materials science, organic and inorganic chemistry, biochem- istry and chemical biology. CWC is freely available for in-house use and is open source (GPL v3) for all other uses. Keywords: ChemDoodle Web Components, Chemical graphics, Animations, Cheminformatics, HTML5, Canvas, JavaScript, WebGL, Structure editor, Structure query Introduction Mobile browsers did support HTML5, which opened How we communicate chemical information is increas- the door to web applications built with only HTML, ingly technology driven. Learning management systems, CSS and JavaScript (JS), such as the ChemDoodle Web virtual classrooms and MOOCs are a few examples where Components. chemistry educators need forward compatible tools for digital natives. Companies that implement emerg- Review ing web technologies can find efficiencies and benefit The ChemDoodle Web Components technology stack from competitive advantages. The first chemical graph- and features ics toolkit for the web, MDL Chime, was introduced in The ChemDoodle Web Components library, released in 1996 [1]. Based on the molecular visualization program 2009, is the first chemistry toolkit for structure viewing RasMol, Chime was developed as a plugin for Netscape and editing that is originally built using only web stand- and later for Internet Explorer and Firefox.