Jlhulm.Pdf (2.103Mb)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Final Copy 2018 09 25 Gaunt
This electronic thesis or dissertation has been downloaded from Explore Bristol Research, http://research-information.bristol.ac.uk Author: Gaunt, Jess Title: A Viral Approach to Translatome Profiling of CA1 Neurons During Associative Recognition Memory Formation General rights Access to the thesis is subject to the Creative Commons Attribution - NonCommercial-No Derivatives 4.0 International Public License. A copy of this may be found at https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode This license sets out your rights and the restrictions that apply to your access to the thesis so it is important you read this before proceeding. Take down policy Some pages of this thesis may have been removed for copyright restrictions prior to having it been deposited in Explore Bristol Research. However, if you have discovered material within the thesis that you consider to be unlawful e.g. breaches of copyright (either yours or that of a third party) or any other law, including but not limited to those relating to patent, trademark, confidentiality, data protection, obscenity, defamation, libel, then please contact [email protected] and include the following information in your message: •Your contact details •Bibliographic details for the item, including a URL •An outline nature of the complaint Your claim will be investigated and, where appropriate, the item in question will be removed from public view as soon as possible. A Viral Approach to Translatome Profiling of CA1 Neurons During Associative Recognition Memory Formation Jessica Ruth Gaunt A dissertation submitted to the University of Bristol in accordance with the requirements for award of the degree of Doctor of Philosophy in the Faculty of Health Sciences, Bristol Medical School. -

Domain of Escherichia Coli 16S Ribosomal RNA Using Site-Directed Photoaffinity Crosslinking
Downloaded from rnajournal.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press RNA (1998), 4:1455–1466+ Cambridge University Press+ Printed in the USA+ Copyright © 1998 RNA Society+ Analysis of the conformation of the 39 major domain of Escherichia coli 16S ribosomal RNA using site-directed photoaffinity crosslinking ALEXANDRE MONTPETIT,1 CATHERINE PAYANT,1 JAMES M. NOLAN,2 and LÉA BRAKIER-GINGRAS1 1Département de Biochimie, Université de Montréal, Montréal, Québec H3T 1J4, Canada 2Department of Biochemistry, Tulane University Medical Center, New Orleans, Louisiana 70112, USA ABSTRACT The 39 major domain of Escherichia coli 16S rRNA, which occupies the head of the small ribosomal subunit, is involved in several functions of the ribosome. We have used a site-specific crosslinking procedure to gain further insights into the higher-order structure of this domain. Circularly permuted RNAs were used to introduce an azi- dophenacyl group at specific positions within the 39 major domain. Crosslinks were generated in a high-ionic strength buffer that has been used for ribosome reconstitution studies and so enables the RNA to adopt a structure recognized by ribosomal proteins. The crosslinking sites were identified by primer extension and confirmed by assessing the mobility of the crosslinked RNA lariats in denaturing polyacrylamide gels. Eight crosslinks were characterized. Among them, one crosslink demonstrates that helix 28 is proximal to the top of helix 34, and two others show that the 1337 region, located in an internal loop at the junction of helices 29, 30, 41, and 42, is proximal to the center of helix 30 and to a segment connecting helix 28 to helix 29. -

Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6
Research Collection Doctoral Thesis Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6 Author(s): Voigts-Hoffmann, Felix Publication Date: 2012 Permanent Link: https://doi.org/10.3929/ethz-a-007303759 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library ETH Zurich Dissertation No. 20189 Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6 A dissertation submitted to ETH ZÜRICH for the degree of Doctor of Sciences (Dr. sc. ETH Zurich) presented by Felix Voigts-Hoffmann MSc Molecular Biotechnology, Universität Heidelberg born April 11, 1981 citizen of Göttingen, Germany accepted on recommendation of Prof. Dr. Nenad Ban (Examiner) Prof. Dr. Raimund Dutzler (Co-examiner) Prof. Dr. Rudolf Glockshuber (Co-examiner) 2012 blank page ii Summary Ribosomes are large complexes of several ribosomal RNAs and dozens of proteins, which catalyze the synthesis of proteins according to the sequence encoded in messenger RNA. Over the last decade, prokaryotic ribosome structures have provided the basis for a mechanistic understanding of protein synthesis. While the core functional centers are conserved in all kingdoms, eukaryotic ribosomes are much larger than archaeal or bacterial ribosomes. Eukaryotic ribosomal rRNA and proteins contain extensions or insertions to the prokaryotic core, and many eukaryotic proteins do not have prokaryotic counterparts. Furthermore, translation regulation and ribosome biogenesis is much more complex in eukaryotes, and defects in components of the translation machinery are associated with human diseases and cancer. -

Supplementary Figure 1
Supplementary Table 1 siRNA Oligonucleotide Sequences not Used for IGFBP-3 Knockdown siRNA Sequence nucleotides Source GCUACAAAGUUGACUACGA 686-704 ON-TARGET Plus SMART pool sequences GAAAUGCUAGUGAGUCGGA 536-554 ON-TARGET Plus SMART pool sequences GCACAGAUACCCAGAACUU 713-731 ON-TARGET Plus SMART pool sequences GAAUAUGGUCCCUGCCGUA 757-775 ON-TARGET Plus SMART pool sequences UAUCGAGAAUAGGAAAACC 1427-1445 siDESIGN center GCAGCCUCUCCCAGGCUACA 940-958 siDESIGN center GCAUAAGCUCUUUAAAGGCA 1895-1913 siDESIGN center UGCCUGGAUUCCACAGCUU 44-62 siDESIGN center AAGCAGCGTGCCCCGGUUG 106-124 siDESIGN center AAAGGCAAAGCUUUAUUUU 1908-1926 siDESIGN center Oligonucleotide sequences used for siRNA oligonucleotides tested to induce IGFBP-3 knockdown. Sequences 1-4 were from ON-TARGET Plus SMART pool sequences (Cat. # L-004777-00-0005, Dharmacon, Lafayette, CO). Sequences 5-10 were generated in our laboratory using the siDESIGN center from the Dharmacon website (www.dharmacon.com) by inputting the Genbank accession number NM_000598 (IGFBP-3). Supplementary Table 2 Transcripts Activated by NKX3.1 in PC-3 Cells PC-3 cells were stably transfected with the pcDNA3.1 empty vector or NKX3.1 expression vector and mRNA from two clones of each cell type was isolated for microarray analysis on the Affymetrix U-133 expression array. Analyses of results from each pair of clones of the same genotype that did not match up were discarded to ensure clonal variation was not a factor. 984 genes were found to be up- or down-regulated more that 1.4 fold in the NKX3.1 expressing PC-3 cells, in comparison to the PC-3 control cells. The 6th and 9th most activated probe sets were for human growth hormone-dependent insulin-like growth factor-binding protein, now known as IGFBP-3. -

Constructing and Analyzing Biological Interaction Networks for Knowledge Discovery
Constructing and Analyzing Biological Interaction Networks for Knowledge Discovery Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Duygu Ucar Graduate Program in Computer Science and Engineering The Ohio State University 2009 Dissertation Committee: Srinivasan Parthasarathy, Advisor Yusu Wang Umit Catalyurek c Copyright by Duygu Ucar 2009 ABSTRACT Many biological datasets can be effectively modeled as interaction networks where nodes represent biological entities of interest such as proteins, genes, or complexes and edges mimic associations among them. The study of these biological network structures can provide insight into many biological questions including the functional characterization of genes and gene products, the characterization of DNA-protein bindings, and the under- standing of regulatory mechanisms. Therefore, the task of constructing biological interac- tion networks from raw data sets and exploiting information from these networks is critical, but is also fraught with challenges. First, the network structure is not always known in a priori; the structure should be inferred from raw and heterogeneous biological data sources. Second, biological networks are noisy (containing unreliable interactions) and incomplete (missing real interactions) which makes the task of extracting useful information difficult. Third, typically these networks have non-trivial topological properties (e.g., uneven degree distribution, small world) that limit the effectiveness of traditional knowledge discovery al- gorithms. Fourth, these networks are usually dynamic and investigation of their dynamics is essential to understand the underlying biological system. In this thesis, we address these issues by presenting a set of computational techniques that we developed to construct and analyze three specific types of biological interaction networks: protein-protein interaction networks, gene co-expression networks, and regulatory networks. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Multifaceted Deregulation of Gene Expression and Protein Synthesis with Age
Multifaceted deregulation of gene expression and protein synthesis with age Aleksandra S. Anisimovaa,b,c,1, Mark B. Meersona,c, Maxim V. Gerashchenkob, Ivan V. Kulakovskiya,d,e,f,2, Sergey E. Dmitrieva,c,d,2, and Vadim N. Gladyshevb,2 aBelozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow 119234, Russia; bDivision of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; cFaculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, Moscow 119234, Russia; dEngelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow 119991, Russia; eVavilov Institute of General Genetics, Russian Academy of Sciences, Moscow 119991, Russia; and fInstitute of Protein Research, Russian Academy of Sciences, Pushchino 142290, Russia Edited by Joseph D. Puglisi, Stanford University School of Medicine, Stanford, CA, and approved May 27, 2020 (received for review January 30, 2020) Protein synthesis represents a major metabolic activity of the cell. RNA polymerase I, eIF2Be, and eEF2, decrease with age in rat However, how it is affected by aging and how this in turn impacts tissues (10). Increased promoter methylation in ribosomal RNA cell function remains largely unexplored. To address this question, genes and decreased ribosomal RNA concentration during aging herein we characterized age-related changes in both the transcrip- were also reported (11). In addition, down-regulation of trans- tome and translatome of mouse tissues over the entire life span. lation with age was confirmed in vivo in the sheep (12) as well as We showed that the transcriptome changes govern those in the in replicatively aged yeast (13). -
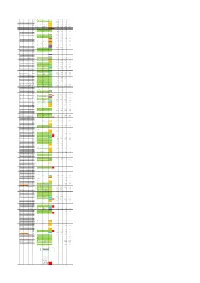
End Strand of Gene Gene Name Gene Function Starnd of Trascript
Genomic TSS strand Gene starnd of Transcipt relatively to TSS Start End Gene name coordinates relatively TSS type Control_fwd TEX_fwd Control_rev TEX_rev of gene function trascript gene group of TSS to ATG ribosomal 93 854 + rps12 + 1 protein S12 1 1 14.0638317 20.7539794 0 0 93 324 exon 1 + 111 rps12 intron 1 1 13.90756687 18.07149224 0.781323982 0.423550599 829 854 exon 2 + 496 rps12 exon2 2 24.22104343 15.24782157 0 0 30S 904 1371 + gene rps7 ribosomal + 2 protein S7 1303 -209 rps7 inter (ndhB) 2 20.47068832 9.035746118 0.625059185 0.847101199 NADH dehydrogen 1512 3535 + gene ndhB (ndh2) + ase subunit 3 2 1696 ndhB exon 1-inter 2 3.594090315 2.964854195 0.468794389 0.282367066 + 2209 ndhB exon 1-inter 2 46.09811492 4.09432246 0.468794389 0.423550599 1512 2237 exon 1 + 2756 ndhB exon 2 -inter 2 43.28534858 4.800240125 0.312529593 0.282367066 2756 3535 exon 2 + 3090 ndhB exon 2 -inter 2 17.50165719 15.95373924 0.312529593 0 + 3192 ndhB exon 2 -inter 2 140.6383167 117.6058831 2.812766334 1.694202397 - 3462 ndhB exon 2 -inter 3 1.406383167 1.129468265 1.406383167 3.67077186 4 3633 3712 + tRNA tRNA-Leu-CAA + 3610 -23 tRNA-Leu 1 77.19480938 84.85130339 0.625059185 0 + 3633 1 tRNA-Leu 1 359.5652963 649.0207016 0.781323982 0 photosyste 3954 4058 + gene psbM m II protein + 5 M 3775 -179 psbM 2 20.47068832 12.00060031 0 0.141183533 + 3954 psbM-0 2 69.22530477 28.37789015 0.156264796 0 hypothetical 4182 5073 + gene ycf66 6 protein 4182 4287 exon 1 4772 5073 exon 2 7 5202 5113 - gene ycf (ORF29) - 5299 orf29 inter 1 0 0 3.125295926 3.67077186 -

(12) United States Patent (10) Patent No.: US 9,476,099 B2 Spinella Et Al
US009476.099B2 (12) United States Patent (10) Patent No.: US 9,476,099 B2 Spinella et al. (45) Date of Patent: Oct. 25, 2016 (54) METHOD FOR DETERMINING FOREIGN PATENT DOCUMENTS SENSTIVITY TO DECTABINE WO WO 2012/031008 A2 3, 2012 TREATMENT WO WO 2012, 11885.6 A1 9, 2012 (71) Applicant: TRUSTEES OF DARTMOUTH COLLEGE, Hanover, NH (US) OTHER PUBLICATIONS (72) Inventors: Michael Spinella, Hanover, NH (US); Tsai et al Cancer Cell. Mar. 20, 2012. 21(3):43.0-446 and Supple Maroun J. Beyrouthy, Lebanon, NH mental pages 1-18. (US) Agilent Technologies. DNA Oligo Microarray Gene Lists and Annotations, available via url: <chem.agilent.com/cag?bSp? gene (73) Assignee: Trustees of Dartmouth College, lists.asp printed on Mar. 1, 2016.* Hanover, NH (US) Abele et al. “The EORTC Early Clinical Trials Cooperative Group Experience with 5-Aza-2'-deoxycytidine (NSC 127716) in Patients (*) Notice: Subject to any disclaimer, the term of this with Colo-rectal, Head and Neck, Renal Carcinomas and Malignant patent is extended or adjusted under 35 Melanomas' European Journal of Cancer and Clinical Oncology U.S.C. 154(b) by 0 days. 1987 23(12): 1921-1924. Adewumi et al. “Characterization of Human Embryonic Stem Cell (21) Appl. No.: 14/416,142 Lines by the International StemCell Initiative” Nature Biotechnol ogy 2007 vol. 25(7):803-816. (22) PCT Filed: Jul. 31, 2013 Al-Hajj et al. "Prospective Identification of Tumorigenic Breast Cancer Cells' Proceedings of the National Academy of Sciences (86). PCT No.: PCT/US2013/052899 2003 100(7):3983-3988 with correction. -

Ribosomal RNA
Ribosomal RNA Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins.[1] Ribosomal RNA Three-dimensional views of the ribosome, showing rRNA in dark blue (small subunit) is the predominant form of RNA found in most cells; it makes and dark red (large subunit). Lighter colors up about 80% of cellular RNA despite never being translated represent ribosomal proteins. into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Contents Structure Assembly Function Subunits and associated ribosomal RNA In prokaryotes In eukaryotes Biosynthesis In eukaryotes Eukaryotic regulation In prokaryotes Prokaryotic regulation Degradation In eukaryotes In prokaryotes Sequence conservation and stability Significance Human genes See also References External links Structure Although the primary structure of rRNA sequences can vary across organisms, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of these rRNA stem-loops allow them to create three-dimensional rRNA structures that are similar across species.[2] Because of these configurations, rRNA can form tight and specific interactions with ribosomal proteins to form ribosomal subunits. These ribosomal proteins contain basic residues (as opposed to acidic residues) and aromatic residues (i.e. -

Supersized Ribosomal RNA Expansion Segments in Asgard Archaea 2 Authors: Petar I
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.25.888164; this version posted June 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Supersized ribosomal RNA expansion segments in Asgard archaea 2 Authors: Petar I. Penev1,2, Sara Fakhretaha-Aval1,3, Vaishnavi J. Patel4, Jamie J. Cannone4, 3 Robin R. Gutell4, Anton S. Petrov1,3*, Loren Dean Williams1,2,3*, Jennifer B. Glass1,2,5* 4 Affiliations: 1NASA Center for the Origin of Life, Georgia Institute of Technology, Atlanta, GA 30332-0400, USA 2School of Biological Sciences, Georgia Institute of Technology, North Avenue, Atlanta, GA 30332, USA 3School of Chemistry and Biochemistry, Georgia Institute of Technology, 901 Atlantic Dr, Atlanta, GA 30332, USA 5 4Department of Integrative Biology, The University of Texas at Austin, 2415 Speedway #C0930, 6 Austin, TX 78712, USA 7 5School of Earth and Atmospheric Sciences, Georgia Institute of Technology, 311 Ferst Dr, 8 Atlanta, GA 30332, USA 9 *Correspondence to: [email protected]; [email protected]; 10 [email protected] 11 1 bioRxiv preprint doi: https://doi.org/10.1101/2019.12.25.888164; this version posted June 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Dissertation
Dissertation submitted to the Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg, Germany for the degree of Doctor of Natural Sciences Presented by Dipl. Biochemist Birgit Rogell born in Speyer (Germany) nd Oral-examination: 2 November 2016 Exploring the biology of RNPs: specific capture of RNPs using antisense locked nucleic acids Referees: 1. Dr. Anne Ephrussi 2. Prof. Dr. Ralf Bartenschlager Abstract Abstract RNA-binding proteins (RBPs) are central players in cell biology and respond to a multitude of cellular cues and environmental stimuli. Identification of RBPs associated with specific transcripts in a cell is a challenging task; and the available strategies to purify specific transcripts and their bound proteome face numerous limitations. Thus, methods to determine the composition of proteins on a given RNA are required to further understand the regulation and biological function of any given RNA. Therefore, the focus of my PhD project was to develop a highly specific and selective method, “specific Ribonucleoprotein (RNP) capture”, to isolate a specific RNA species together with its bound proteome. Following irradiation with ultraviolet (UV) light that creates a covalent bond between RNA and protein, RNAs are captured using short LNA (locked nucleic acid)/DNA mixmer antisense probes coupled to a solid support.The proteins covalently linked to the isolated RNA are then identified by quantitative mass spectrometry. First, I successfully established the method for this application in vitro. Mass spectrometry data revealed that the protein Sister of Sex lethal (Ssx) has similar binding preferences to a mRNA derived from male-specific lethal (msl)2 mRNA as its paralog Sxl in Drosophila melanogaster embryo extracts.