Download Image
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mothers in Science
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -
BSCB Newsletter 2017D
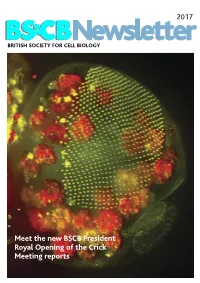
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -

Female Fellows of the Royal Society
Female Fellows of the Royal Society Professor Jan Anderson FRS [1996] Professor Ruth Lynden-Bell FRS [2006] Professor Judith Armitage FRS [2013] Dr Mary Lyon FRS [1973] Professor Frances Ashcroft FMedSci FRS [1999] Professor Georgina Mace CBE FRS [2002] Professor Gillian Bates FMedSci FRS [2007] Professor Trudy Mackay FRS [2006] Professor Jean Beggs CBE FRS [1998] Professor Enid MacRobbie FRS [1991] Dame Jocelyn Bell Burnell DBE FRS [2003] Dr Philippa Marrack FMedSci FRS [1997] Dame Valerie Beral DBE FMedSci FRS [2006] Professor Dusa McDuff FRS [1994] Dr Mariann Bienz FMedSci FRS [2003] Professor Angela McLean FRS [2009] Professor Elizabeth Blackburn AC FRS [1992] Professor Anne Mills FMedSci FRS [2013] Professor Andrea Brand FMedSci FRS [2010] Professor Brenda Milner CC FRS [1979] Professor Eleanor Burbidge FRS [1964] Dr Anne O'Garra FMedSci FRS [2008] Professor Eleanor Campbell FRS [2010] Dame Bridget Ogilvie AC DBE FMedSci FRS [2003] Professor Doreen Cantrell FMedSci FRS [2011] Baroness Onora O'Neill * CBE FBA FMedSci FRS [2007] Professor Lorna Casselton CBE FRS [1999] Dame Linda Partridge DBE FMedSci FRS [1996] Professor Deborah Charlesworth FRS [2005] Dr Barbara Pearse FRS [1988] Professor Jennifer Clack FRS [2009] Professor Fiona Powrie FRS [2011] Professor Nicola Clayton FRS [2010] Professor Susan Rees FRS [2002] Professor Suzanne Cory AC FRS [1992] Professor Daniela Rhodes FRS [2007] Dame Kay Davies DBE FMedSci FRS [2003] Professor Elizabeth Robertson FRS [2003] Professor Caroline Dean OBE FRS [2004] Dame Carol Robinson DBE FMedSci -

The Control of Shoot Branching: an Example of Plant Information Processing
Plant, Cell and Environment (2009) 32, 694–703 doi: 10.1111/j.1365-3040.2009.01930.x The control of shoot branching: an example of plant information processing OTTOLINE LEYSER Department of Biology, Area 11, University of York, York YO10 5YW, UK ABSTRACT the apical–basal axis defined by the establishment of the shoot apical meristem at one end, and the root apical mer- Throughout their life cycle, plants adjust their body plan istem at the other. Post-embryonically, the meristems give to suit the environmental conditions in which they are rise to the entire shoot and root systems, respectively. Fur- growing. A good example of this is in the regulation of thermore, the tissues they establish can produce secondary shoot branching. Axillary meristems laid down in each leaf meristems, which if activated can produce entirely new formed from the primary shoot apical meristem can remain axes of growth with the same developmental potential as dormant, or activate to produce a branch. The decision the primary root or shoot from which they were derived. whether to activate an axillary meristem involves the assess- Thus, the body plan of a plant is determined continuously ment of a wide range of external environmental, internal throughout its life cycle, allowing it to be exquisitely envi- physiological and developmental factors. Much of this infor- ronmentally responsive. Plants can alter their body plan to mation is conveyed to the axillary meristem via a network suit their environment, and thus the environmentally regu- of interacting hormonal signals that can integrate inputs lated development of new growth axes is functionally from diverse sources, combining multiple local signals to equivalent to environmentally regulated animal behav- generate a rich source of systemically transmitted informa- iours. -

Annual Report 2015
Annual Report 2015 Clare College Cambridge Contents Master’s Introduction .................................................................... 3 Teaching and Research .............................................................. 4–5 Selected Publications by Clare Fellows ....................................... 6–9 College Life ........................................................................... 10–12 Access & Outreach ..................................................................... 13 Financial Report ..................................................................... 14–15 Development ....................................................................... 16–17 List of Master & Fellows............................................................... 18 Captions ..................................................................................... 19 2 Master’s Introduction The past year has been full of many introductions for me two new CDs. They are fantastic ambassadors for the College and met some of our alumni in New as a relatively new Master - to student life in its various York - looking ahead; in 2016 they travel to Hong Kong, Malaysia and Singapore for more concerts. forms, to colleagues, and to our alumni at various events at home and abroad. The College continues to be in Last year I mentioned the need to refurbish Old Court, and this is still very much our priority. The good form, and weathering various new initiatives from Fellowship has just approved - in principle - plans which are now before Historic England, -

Selective Auxin Agonists Induce Specific AUX/IAA Protein Degradation to Modulate Plant Development
Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development Thomas Vaina,1,2, Sara Raggia,1, Noel Ferrob, Deepak Kumar Barangea,c, Martin Kiefferd, Qian Maa, Siamsa M. Doylea, Mattias Thelandere, Barbora Parízkováf,g, Ondrej Nováka,f,g, Alexandre Ismailh, Per-Anders Enquistc, Adeline Rigala, Małgorzata Łangowskaa, Sigurd Ramans Harboroughd, Yi Zhangi, Karin Ljunga, Judy Callisj, Fredrik Almqvistc, Stefan Kepinskid, Mark Estellei, Laurens Pauwelsk,l, and Stéphanie Roberta,3 aDepartment of Forest Genetics and Plant Physiology, Umeå Plant Science Centre, Swedish University of Agricultural Sciences, SE-901 83 Umeå, Sweden; bInstitute of Physical and Theoretical Chemistry, University of Bonn, 53121 Bonn, Germany; cLaboratories for Chemical Biology Umeå, Chemical Biology Consortium Sweden, Department of Chemistry, Umeå University, SE-901 87 Umeå, Sweden; dCentre for Plant Sciences, University of Leeds, LS2 9JT Leeds, United Kingdom; eDepartment of Plant Biology, Swedish University of Agricultural Sciences, The Linnean Centre for Plant Biology in Uppsala, SE-75007 Uppsala, Sweden; fLaboratory of Growth Regulators, Institute of Experimental Botany, The Czech Academy of Sciences, CZ-78371 Olomouc, Czech Republic; gLaboratory of Growth Regulators, Faculty of Science, Palacký University, CZ-78371 Olomouc, Czech Republic; hSup’Biotech, IONIS Education Group, 94800 Villejuif, France; iSection of Cell and Developmental Biology, University of California, San Diego, La Jolla, CA 92093-0116; jDepartment of Molecular and Cellular Biology, University of California, Davis, CA 95616; kDepartment of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Ghent, Belgium; and lCenter for Plant Systems Biology, Vlaams Instituut voor Biotechnologie, 9052 Ghent, Belgium Edited by Ottoline Leyser, University of Cambridge, Cambridge, United Kingdom, and approved February 6, 2019 (received for review May 25, 2018) Auxin phytohormones control most aspects of plant development transcriptional response. -

Engage Festival 2020 Engagement, Innovation and Change
Engage Festival 2020 Engagement, Innovation and Change National Co-ordinating Centre for Public Engagement #Engage2020 @NCCPE 01 NCCPE Engage Festival 2020 Introduction We’re really looking forward to welcoming you to the Engage Festival 2020 The Engage Festival is a week-long extravaganza of over 60 live events inviting you to explore and reimagine the role and future of public engagement, as COVID-19 redefines our world. Running from Monday 30th November – Friday 4th December, we have developed a rich programme of thought-provoking talks, interactive workshops, immersive experiences, and lots of creative ways for people to connect and network. Everyone’s experience of our current context is different, and we have timetabled our content to be suitable for a range of availabilities and engagement preferences. We invite people to curate their own unique engagement experience, picking and choosing from the options on offer – and ensuring the conference is tailored to you. This draft programme outlines the sessions on offer during the festival, including the platforms they’ll be hosted on. Each workshop lead is listed, and they will all be joined in their sessions by contributors from the wider sector. The Engage Festival will also be broadcasting live every day! So why not tune in to find out what has been happening during the day, and hear delegates, workshops leaders and our festival reporters reflecting on the festival themes. It provide a great way to hear the latest news, and to catch up on what you have missed. Facilitated by Jamie Gallagher, the daily review livestream will broadcast every afternoon. -

REPORTER S P E C I a L No 6 T U E S D Ay 24 a P R I L 2018 Vol Cxlviii
CAMBRIDGE UNIVERSITY REPORTER S PECIAL NO 6 T UE S D AY 24 A PRIL 2018 VOL CXLVIII MEMBERS OF UNIVERSITY BODIES REPRESENTATIVES OF THE UNIVERSITY (‘OFFICERS NUMBER’, PARTS II AND III) PUBLISHED BY AUTHORITY [SPECIAL NO. 6 MEMBERS OF UNIVERSITY BODIES REPRESENTATIVES OF THE UNIVERSITY P ART II: M E mb ER S OF U N I V ER S I T Y B ODIE S Nominating and appointing bodies: abbreviations 1 Faculty Boards and Degree Committees 18 Septemviri, Discipline Committee, University Tribunal 1 Committees 26 Discipline Board 2 Trustees, Managers, Awarders, of Funds, Council, Audit Committee, Finance Committee 2 Scholarships, Studentships, Prizes, etc. 32 General Board of the Faculties 2 Representatives of the Colleges for Election of Other Committees of the Central Bodies 2 Members of the Finance Committee 51 Boards of Electors to Professorships 6 P ART III: R EPRE S E ntAT I V E S OF th E U N I V ER S I T Y Advisory Committees for Elections to Professorships 7 Boards of Electors to offices other than Professorships 7 1. Representative Governors, etc. 52 Syndicates 8 2. Representative Trustees Associated with the Boards 9 University 53 Councils of the Schools 11 3. Cambridge Enterprise Ltd: Board of Directors 53 Appointments Committees 12 NOTICE BY thE EDITOR Following the publication of Part I of the Officers Number in February 2018, this issue of Part II (Members of University Bodies) and Part III (Representatives of the University) includes data received up to 13 April 2018. The next update of Part I (University Officers) will be published in Lent Term 2019 and an update to Parts II and III will follow shortly thereafter. -

Network Trade-Offs and Homeostasis in Arabidopsis Shoot Architectures
RESEARCH ARTICLE Network trade-offs and homeostasis in Arabidopsis shoot architectures 1 1 2 2 Adam ConnID , Arjun ChandrasekharID , Martin van Rongen , Ottoline LeyserID , 3 1 Joanne ChoryID , Saket NavlakhaID * 1 Integrative Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California, United States of America, 2 Sainsbury Laboratory, University of Cambridge, Cambridge, United Kingdom, 3 Howard Hughes Medical Institute and Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California, United States of America a1111111111 * [email protected] a1111111111 a1111111111 a1111111111 a1111111111 Abstract Understanding the optimization objectives that shape shoot architectures remains a critical problem in plant biology. Here, we performed 3D scanning of 152 Arabidopsis shoot archi- tectures, including wildtype and 10 mutant strains, and we uncovered a design principle that OPEN ACCESS describes how architectures make trade-offs between competing objectives. First, we used Citation: Conn A, Chandrasekhar A, Rongen Mv, graph-theoretic analysis to show that Arabidopsis shoot architectures strike a Pareto optimal Leyser O, Chory J, Navlakha S (2019) Network that can be captured as maximizing performance in transporting nutrients and minimizing trade-offs and homeostasis in Arabidopsis shoot costs in building the architecture. Second, we identify small sets of genes that can be architectures. PLoS Comput Biol 15(9): e1007325. https://doi.org/10.1371/journal.pcbi.1007325 mutated to shift the weight prioritizing one objective over the other. Third, we show that this prioritization weight feature is significantly less variable across replicates of the same geno- Editor: Giovanni Stracquadanio, University of Oxford, UNITED KINGDOM type compared to other common plant traits (e.g., number of rosette leaves, total volume occupied). -
2011-06-15-Mothers-In-Science.Pdf
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -

EVENT PROGRAMME WELCOME UKRIO Dear Friends and Colleagues
UKRIO UKRIO ANNUAL CONFERENCE 2021 EVENT PROGRAMME WELCOME UKRIO Dear friends and colleagues, I hope you and your loved ones are well in these challenging and changing times. The research community, like every part of society, has been greatly affected by the pandemic. Alongside the immense impact on health and wellbeing, how we work has had to change, sometimes radically. Existing pressures on researchers have heightened and new challenges have arisen. Following the changes imposed on the research community out of necessity, there has been increased discussion of our working practices and the pressures we face trying to ensure that our research is the best that it can be. How can we change things for the better? What practices should we retain and what might we do differently? I’m delighted that you are joining us for this year’s UKRIO conference. Our speakers will explore issues of good research practice in this changing world that we are living in, sharing lessons learned and also looking to the future. I hope you enjoy our event and I’m sure the fascinating talks will generate loads of interesting discussion. Our online events programme is allowing us to reach more people than ever and we’ll be continuing that in 2021 and beyond, with regular webinars and other virtual events. Please visit our website for more information on the work of our charity, including information on how to volunteer with us and how institutions can subscribe to UKRIO. James Parry Chief Executive, UK Research Integrity Office UKRIO ANNUAL CONFERENCE 2021 2 EVENT OVERVIEW Building on the successes of UKRIO’s monthly research integrity webinars, we This event will allow for interaction and encourages participants to share their have taken our annual conference entirely online. -

2013 Faseb Science Research Conferences Advisory Committee Meeting Topic for Consideration
Proposal #: 15-16 2013 FASEB SCIENCE RESEARCH CONFERENCES ADVISORY COMMITTEE MEETING TOPIC FOR CONSIDERATION TOPIC NAME: MECHANISMS OF PLANT DEVELOPMENT PREVIOUS TITLE: Mechanisms in Plant Development SUBMITTED BY: Dominique Bergmann, HHMI and Department of Biology, Stanford University Rüdiger Simon, Institute of Developmental Genetics, Heinrich-Heine University YEAR REQUESTED FOR 2015 SCHEDULING: SITE REQUESTS: 1. Saxtons River, VT 2. Keystone, CO 3. Steamboat Springs, CO DATE REQUESTS: 1. Aug 1-6, 2015 2. Aug 8-14, 2015 3. Aug 16-22, 2015 YEAR(S) CONFERENCE 1987, 1989, 1992, 1994, 1996, 1998, 2000, 2002, 2004, 2006, 2008, 2010, 2013 HAS BEEN HELD: NOTES: To the best of our knowledge, there is not a direct conflict with any other FASEB SRC or other society or industry meeting. Dear Colleague, We invite you to submit a proposal for a future FASEB Science Research Conference Series (SRC). Since 1982, FASEB has worked hand-in-hand with scientists to organize conferences for experimental biologists. The Conferences are divided up into small groups, who meet intimately and without distractions to explore new approaches to research areas undergoing rapid scientific change. FASEB supports over 35 SRCs each year. Site preferences for 2015, 2016, and 2017 are Big Sky, MT, Chicago, IL, Saxtons River, VT, Snowmass, CO, Steamboat Springs, CO, Nassau, Bahamas, Keystone, CO, Liverpool, England, Palm Beach, FL Palm Springs, CA, Reno/Las Vegas, NV, and Lisbon, Portugal Site preference selection will be prioritized by site availability, history of conference success and registration fee factors. Additionally, FASEB welcomes new site suggestions that are conducive to conference requirements. Please review the attached information and feel free to contact the SRC Office should you have any questions or need guidance in preparing your proposal.