Scientific Report 2003
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mothers in Science
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -
BSCB Newsletter 2017D
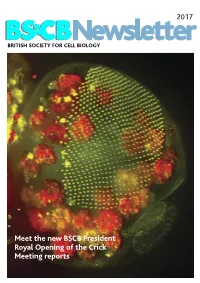
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -
![Winter Memory Throughout the Plant Kingdom: Different Paths to Flowering1[OPEN]](https://docslib.b-cdn.net/cover/2772/winter-memory-throughout-the-plant-kingdom-different-paths-to-flowering1-open-252772.webp)
Winter Memory Throughout the Plant Kingdom: Different Paths to Flowering1[OPEN]
Update on Vernalization Pathways Winter Memory throughout the Plant Kingdom: Different Paths to Flowering1[OPEN] Frédéric Bouché, Daniel P. Woods, and Richard M. Amasino* Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706 (F.B., D.P.W., R.M.A.); and United States Department of Energy Great Lakes Bioenergy Research Center, Madison, Wisconsin 53726 (D.P.W., R.M.A.) ORCID IDs: 0000-0002-8017-0071 (F.B.); 0000-0002-1498-5707 (D.P.W.); 0000-0003-3068-5402 (R.M.A.). Plants have evolved a variety of mechanisms to syn- developmental program that prevents flowering in young chronize flowering with their environment to optimize seedlings and promotes the transition to reproductive de- reproductive success. Many species flower in spring when velopmentinolderplants(e.g.Yuetal.,2015).Inmany the photoperiod increases and the ambient tempera- species adapted to temperate climates, the perception tures become warmer. Winter annuals and biennials have of seasonal changes also involves the acquisition of the evolved repression mechanisms that prevent the transition competence to flower in response to an extended cold to reproductive development in the fall. These repressive period, a process referred to as vernalization (e.g. Chouard, processes can be overcome by the prolonged cold of 1960; Preston and Sandve, 2013; Fig. 1A). In addition, some winter through a process known as vernalization. The species acquire floral competence when exposed to the memory of the past winter is sometimes stored by epige- shorter photoperiod of winter (Purvis and Gregory, 1937; netic chromatin remodeling processes that provide com- Wellensiek, 1985), but the molecular mechanisms control- petence to flower, and plants usually require additional ling the so-called “short-day vernalization” are still un- inductive signals to flower in spring. -

Focus on Ghent University
FOCUS ON GHENT UNIVERSITY 4 FOREWORD In ‘Focus on Ghent University’ we proudly present our higher education institution. Ghent University is an enterprising university with an international appeal. Our motto is ‘Dare to think’: we encourage students and staff members to adopt a critical approach. Pluralism and social engagement are the historic cornerstones of our philosophy. Diversity, participation and independence are common denominators in our policies. This publication describes our key tasks: education, scientific research and public service delivery. The university is a unique biotope of academic education and scientific research, pillars that are inseparably linked with one another. The cross-fertilization between education and research is vital if we want to continue to play a leading innovative role. In ‘Focus on Ghent University’ we also concentrate on the close links between Ghent University and the city and region. Together with our local and (inter)national partners, we play a major role as a catalyst of innovation in all areas. Finally, in this publication we take a look at student facilities and the role of our university as the largest employer in East Flanders. FOCUS ON GHENT UNIVERSITY FOREWORD 5 GHENT UNIVERSITY IS … GHENT AREA DARE TO THINK CAMPUS KORTRIJK ‘Dare to Think’ is the credo of Ghent University: critical and independent minds study, carry out research and work at Ghent University. CAMPUS OSTEND The phrase is the literal translation of Sapere aude that was originally used by the Roman poet Horace and was also the main idea of enlightened critical philosophers such as Immanuel Kant. PLURALISM AND PARTICIPATION Ghent University is a socially engaged pluralistic university that is open to all students and staff irrespective of their ideological, political, cultural and social background. -

Etude Sur L'origine Et L'évolution Des Variations Florales Chez Delphinium L. (Ranunculaceae) À Travers La Morphologie, L'anatomie Et La Tératologie
Etude sur l'origine et l'évolution des variations florales chez Delphinium L. (Ranunculaceae) à travers la morphologie, l'anatomie et la tératologie : 2019SACLS126 : NNT Thèse de doctorat de l'Université Paris-Saclay préparée à l'Université Paris-Sud ED n°567 : Sciences du végétal : du gène à l'écosystème (SDV) Spécialité de doctorat : Biologie Thèse présentée et soutenue à Paris, le 29/05/2019, par Felipe Espinosa Moreno Composition du Jury : Bernard Riera Chargé de Recherche, CNRS (MECADEV) Rapporteur Julien Bachelier Professeur, Freie Universität Berlin (DCPS) Rapporteur Catherine Damerval Directrice de Recherche, CNRS (Génétique Quantitative et Evolution Le Moulon) Présidente Dario De Franceschi Maître de Conférences, Muséum national d'Histoire naturelle (CR2P) Examinateur Sophie Nadot Professeure, Université Paris-Sud (ESE) Directrice de thèse Florian Jabbour Maître de conférences, Muséum national d'Histoire naturelle (ISYEB) Invité Etude sur l'origine et l'évolution des variations florales chez Delphinium L. (Ranunculaceae) à travers la morphologie, l'anatomie et la tératologie Remerciements Ce manuscrit présente le travail de doctorat que j'ai réalisé entre les années 2016 et 2019 au sein de l'Ecole doctorale Sciences du végétale: du gène à l'écosystème, à l'Université Paris-Saclay Paris-Sud et au Muséum national d'Histoire naturelle de Paris. Même si sa réalisation a impliqué un investissement personnel énorme, celui-ci a eu tout son sens uniquement et grâce à l'encadrement, le soutien et l'accompagnement de nombreuses personnes que je remercie de la façon la plus sincère. Je remercie très spécialement Florian Jabbour et Sophie Nadot, mes directeurs de thèse. -

Studies in <I>Erysiphales</I> Anamorphs (4): Species on <I>Hydrangeaceae</I> and <I>Papaveraceae&L
ISSN (print) 0093-4666 © 2011. Mycotaxon, Ltd. ISSN (online) 2154-8889 MYCOTAXON Volume 115, pp. 287–301 January–March 2011 doi: 10.5248/115.287 Studies in Erysiphales anamorphs (4): species on Hydrangeaceae and Papaveraceae Anke Schmidt1 & Markus Scholler2* 1Holunderweg 2 B, D-23568 Lübeck, Germany 2Staatliches Museum für Naturkunde, Erbprinzenstr. 13, D-76133 Karlsruhe, Germany * Correspondence to: [email protected] Abstract — Anamorphic powdery mildews on Hydrangeaceae and Papaveraceae in Germany are revised. Species are documented in detail including line drawings, photomicrographs, and identification keys. On Papaveraceae three species are accepted, specifically Erysiphe macleayae on Chelidonium majus and Macleaya cordata, E. cruciferarum on Eschscholzia californica, and Oidium sp. (an unknown species previously assigned to E. cruciferarum) on Pseudofumaria lutea. Species on Hydrangeaceae are Oidium hortensiae on Hydrangea macrophylla and E. deutziae on Deutzia cf. scabra and Philadelphus cf. coronarius. The fungus on the latter host plant was previously assigned to O. hortensiae. Erysiphe deutziae, E. macleayae, and Oidium hortensiae are introduced species. Key words — conidial germination, morphology, neomycete Introduction In Germany, there are three species of Erysiphales reported on Papaveraceae (Erysiphe cruciferarum, Erysiphe cf. macleayae, Golovinomyces orontii (Castagne) Heluta); and two (Erysiphe deutziae, Oidium hortensiae) on Hydrangeaceae (Braun 1995, Jage et. al. 2010). The following is a revision of anamorphs on certain host plants of Papaveraceae/Hydrangeaceae (Chelidonium, Deutzia, Hydrangea, Macleaya, Meconopsis, Philadelphus and Pseudofumaria) for which the host/pathogen affiliations have been doubtful. Materials & methods Both fresh and dried structures were examined in tap water mounts with light microscopy using Olympus BH 2 and Zeiss Axioskop 2 Plus. -

To What Extent Can the Phenotypic Differences Between Misopates Orontium and Antirrhinum Majus Be Bridged by Mutagenesis?
Bioremediation, Biodiversity and Bioavailability ©2007 Global Science Books Biodiversity and Dollo’s Law: To What Extent can the Phenotypic Differences between Misopates orontium and Antirrhinum majus be Bridged by Mutagenesis? Wolf-Ekkehard Lönnig1* • Kurt Stüber1 • Heinz Saedler1 • Jeong Hee Kim1 1 Max-Planck-Institute for Plant Breeding Reseach, Carl-von-Linné-Weg 10, 50829 Cologne, Federal Republic of Germany Corresponding author : * [email protected] ABSTRACT According to Dollo’s law, evolution is irreversible. Yet, of the eight derived features essentially distinguishing Misopates orontium from its closely related Antirrhinum majus, five differences have phenotypically been clearly diminished or fully overcome by mutant genes, so that Misopates orontium outwardly approaches, meets or even overlaps the features of Antirrhinum majus or vice versa (aspects of the life cycle, leaf form, flower size, flower colour and mode of fertilization). However, to date the morphological key distinguishing feature between the two genera, the strongly elongated sepals in Misopates (itself a feature being at odds with Dollo’s law), could not be reduced to that of the length of Antirrhinum nor could the development of the short Antirrhinum sepals be extended to that of the length of Misopates, in spite of extensive mutagenesis programmes with both species (agreeing with Dollo’s law as to the stasis of this difference). Also, the long sepal character strongly dominated almost all homeotic Misopates mutants. After a general discussion of Dollo’s -

Memory of the Vernalized State in Plants Including the Model Grass Brachypodium Distachyon
PERSPECTIVE ARTICLE published: 25 March 2014 doi: 10.3389/fpls.2014.00099 Memory of the vernalized state in plants including the model grass Brachypodium distachyon Daniel P.Woods1,2,3 ,Thomas S. Ream1,2 † and Richard M. Amasino1,2 * 1 Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, USA 2 U.S. Department of Energy–Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI, USA 3 Laboratory of Genetics, University of Wisconsin-Madison, Madison, WI, USA Edited by: Plant species that have a vernalization requirement exhibit variation in the ability to George Coupland, Max Planck “remember” winter – i.e., variation in the stability of the vernalized state. Studies in Society, Germany Arabidopsis have demonstrated that molecular memory involves changes in the chromatin Reviewed by: state and expression of the flowering repressor FLOWERING LOCUS C, and have revealed Paula Casati, Centro de Estudios Fotosinteticos – Consejo Nacional de that single-gene differences can have large effects on the stability of the vernalized state. In Investigaciones Científicas y Técnicas, the perennial Arabidopsis relative Arabis alpina, the lack of memory of winter is critical for its Argentina perennial life history. Our studies of flowering behavior in the model grass Brachypodium Iain Robert Searle, The University of Adelaide, Australia distachyon reveal extensive variation in the vernalization requirement, and studies of a particular Brachypodium accession that has a qualitative requirement for both cold exposure *Correspondence: Richard M. Amasino, Department of and inductive day length to flower reveal that Brachypodium can exhibit a highly stable Biochemistry, University of vernalized state. Wisconsin-Madison, 433 Babcock Drive, Madison, WI 53706-1544, USA Keywords: vernalization, flowering, Brachypodium, epigenetics, life history e-mail: [email protected] †Present address: Thomas S. -

Cataleg Biodiversitat Albufera Mallorca 1998
Edita: Conselleria de Medi Ambient Direcció General de Biodiversitat Dibuix de la coberta: Joan Miquel Bennàssar Impressió: Gràfiques Son Espanyolet Diposit Legal: PM 1992-2002 Pròleg Aquesta obra que teniu a les mans és el producte d’un llarg procés de recerca i d’investiga- ció. És el resultat de més de 12 anys de treballs que han duit a terme els científics del grup s‘Albufera-IBG i molts d‘altres experts i aficionats que han aportat l’esforç i els coneixe- ments per completar aquesta visió global de la biodiversitat de s‘Albufera de Mallorca. Un cos de coneixements que s’ha anat formant pas a pas, com és característic de tot procés cien- tífic. La Conselleria de Medi Ambient és ben conscient que la conservació del patrimoni natural no es pot fer sense els estudis que ens han de permetre conèixer quins són els recursos sota la nostra responsabilitat i quin valor tenen. De cada vegada el gran públic coneix més quines són les espècies amenaçades i les accions per conservar-les troben ressò en els mitjans de comunicació i en la sensibilitat de la socie- tat en general. Però cal demanar-se: estam fent tot allò que és necessari per garantir la con- servació del patrimoni natural per a les generacions pròximes? Són suficients els coneixe- ments actuals d’aquest patrimoni? Quines espècies o quines poblacions són les més valuo- ses? Aquest llibre ajudarà a respondre algunes d‘aquestes preguntes. És un compromís entre la visió del científic, que sempre desitja aprofundir en l’estudi, i els gestors, que demanen amb urgència unes eines adequades per planificar les accions de conservació i avaluar-les. -

Historical Biogeography of Endemic Seed Plant Genera in the Caribbean: Did Gaarlandia Play a Role?
Received: 18 May 2017 | Revised: 11 September 2017 | Accepted: 14 September 2017 DOI: 10.1002/ece3.3521 ORIGINAL RESEARCH Historical Biogeography of endemic seed plant genera in the Caribbean: Did GAARlandia play a role? María Esther Nieto-Blázquez1 | Alexandre Antonelli2,3,4 | Julissa Roncal1 1Department of Biology, Memorial University of Newfoundland, St. John’s, NL, Canada Abstract 2Department of Biological and Environmental The Caribbean archipelago is a region with an extremely complex geological history Sciences, University of Göteborg, Göteborg, and an outstanding plant diversity with high levels of endemism. The aim of this study Sweden was to better understand the historical assembly and evolution of endemic seed plant 3Gothenburg Botanical Garden, Göteborg, Sweden genera in the Caribbean, by first determining divergence times of endemic genera to 4Gothenburg Global Biodiversity Centre, test whether the hypothesized Greater Antilles and Aves Ridge (GAARlandia) land Göteborg, Sweden bridge played a role in the archipelago colonization and second by testing South Correspondence America as the main colonization source as expected by the position of landmasses María Esther Nieto-Blázquez, Biology Department, Memorial University of and recent evidence of an asymmetrical biotic interchange. We reconstructed a dated Newfoundland, St. John’s, NL, Canada. molecular phylogenetic tree for 625 seed plants including 32 Caribbean endemic gen- Emails: [email protected]; menietoblazquez@ gmail.com era using Bayesian inference and ten calibrations. To estimate the geographic range of the ancestors of endemic genera, we performed a model selection between a null and Funding information NSERC-Discovery grant, Grant/Award two complex biogeographic models that included timeframes based on geological Number: RGPIN-2014-03976; MUN’s information, dispersal probabilities, and directionality among regions. -

Second Contribution to the Vascular Flora of the Sevastopol Area
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Wulfenia Jahr/Year: 2015 Band/Volume: 22 Autor(en)/Author(s): Seregin Alexey P., Yevseyenkow Pavel E., Svirin Sergey A., Fateryga Alexander Artikel/Article: Second contribution to the vascular flora of the Sevastopol area (the Crimea) 33-82 © Landesmuseum für Kärnten; download www.landesmuseum.ktn.gv.at/wulfenia; www.zobodat.at Wulfenia 22 (2015): 33 – 82 Mitteilungen des Kärntner Botanikzentrums Klagenfurt Second contribution to the vascular flora of the Sevastopol area (the Crimea) Alexey P. Seregin, Pavel E. Yevseyenkov, Sergey A. Svirin & Alexander V. Fateryga Summary: We report 323 new vascular plant species for the Sevastopol area, an administrative unit in the south-western Crimea. Records of 204 species are confirmed by herbarium specimens, 60 species have been reported recently in literature and 59 species have been either photographed or recorded in field in 2008 –2014. Seventeen species and nothospecies are new records for the Crimea: Bupleurum veronense, Lemna turionifera, Typha austro-orientalis, Tyrimnus leucographus, × Agrotrigia hajastanica, Arctium × ambiguum, A. × mixtum, Potamogeton × angustifolius, P. × salicifolius (natives and archaeophytes); Bupleurum baldense, Campsis radicans, Clematis orientalis, Corispermum hyssopifolium, Halimodendron halodendron, Sagina apetala, Solidago gigantea, Ulmus pumila (aliens). Recently discovered Calystegia soldanella which was considered to be extinct in the Crimea is the most important confirmation of historical records. The Sevastopol area is one of the most floristically diverse areas of Eastern Europe with 1859 currently known species. Keywords: Crimea, checklist, local flora, taxonomy, new records A checklist of vascular plants recorded in the Sevastopol area was published seven years ago (Seregin 2008). -

THE IRISH RED DATA BOOK 1 Vascular Plants
THE IRISH RED DATA BOOK 1 Vascular Plants T.G.F.Curtis & H.N. McGough Wildlife Service Ireland DUBLIN PUBLISHED BY THE STATIONERY OFFICE 1988 ISBN 0 7076 0032 4 This version of the Red Data Book was scanned from the original book. The original book is A5-format, with 168 pages. Some changes have been made as follows: NOMENCLATURE has been updated, with the name used in the 1988 edition in brackets. Irish Names and family names have also been added. STATUS: There have been three Flora Protection Orders (1980, 1987, 1999) to date. If a species is currently protected (i.e. 1999) this is stated as PROTECTED, if it was previously protected, the year(s) of the relevant orders are given. IUCN categories have been updated as follows: EN to CR, V to EN, R to V. The original (1988) rating is given in brackets thus: “CR (EN)”. This takes account of the fact that a rare plant is not necessarily threatened. The European IUCN rating was given in the original book, here it is changed to the UK IUCN category as given in the 2005 Red Data Book listing. MAPS and APPENDIX have not been reproduced here. ACKNOWLEDGEMENTS We are most grateful to the following for their help in the preparation of the Irish Red Data Book:- Christine Leon, CMC, Kew for writing the Preface to this Red Data Book and for helpful discussions on the European aspects of rare plant conservation; Edwin Wymer, who designed the cover and who, as part of his contract duties in the Wildlife Service, organised the computer applications to the data in an efficient and thorough manner.