Co-Expression Network Approach Reveals Functional Similarities Among Diseases Affecting Human Skeletal Muscle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diseases/Symptoms Reported to Be Associated with Lyme Disease
Diseases/Symptoms Reported to be Associated with Lyme Disease Abdominal pseudo-eventration Abdominal wall weakness Acrodermatitis chronica atrophicans (ACA) Acute Acral Ischemia Acute conduction disorders Acute coronary syndrome Acute exogenous psychosis Acute febrile illness Acute hemiparesis Acute ischaemic pontine stroke Acute meningitis Acute myelo-meningo-radiculitis Acute myelitis Acute pediatric monoarticular arthritis Acute peripheral facial palsy Acute perimyocarditis Acute posterior multifocal placoid pigment epitheliopathy (APMPPE) Acute pyogenic arthritis Acute reversible diffuse conduction system disease Acute septic arthritis Acute severe encephalitis Acute transitory auriculoventricular block Acute transverse myelitis Acute urinary retention Acquired Immune Deficiency Syndrome (AIDS) Algodystrophy Allergic conditions Allergic conjunctivitis Alopecia Alzheimer’s Disease Amyotrophic lateral sclerosis (ALS - Lou Gehrig's Disease) Amyotrophy Anamnesis Anetoderma Anorexia nervosa Anterior optic neuropathy Antepartum fever Anxiety Arrhythmia Arthralgia Arthritis Asymmetrical hearing loss Ataxic sensory neuropathy Atraumatic spontaneous hemarthrosis Atrioventricular block Attention Deficit Disorder (ADD) Attention Deficit Hyperactivity Disorder (ADHD) Back pain without radiculitis Bannwarth’s Syndrome Behcet's disease Bell’s Palsy Benign cutaneous lymphocytoma Benign lymphocytic infiltration (Jessner-Kanof) Bilateral acute confluent disseminated choroiditis Bilateral carpal tunnel syndrome Bilateral facial nerve palsy Bilateral -

ICD-10-CM TABULAR LIST of DISEASES and INJURIES 2018 Addenda No Change Chapter 1 No Change Certain Infectious and Parasitic Diseases (A00-B99)
ICD-10-CM TABULAR LIST of DISEASES and INJURIES 2018 Addenda No Change Chapter 1 No Change Certain infectious and parasitic diseases (A00-B99) No Change Intestinal infectious diseases (A00-A09) No Change A04 Other bacterial intestinal infections No Change A04.7 Enterocolitis due to Clostridium difficile Add A04.71 Enterocolitis due to Clostridium difficile, recurrent Add A04.72 Enterocolitis due to Clostridium difficile, not specified as recurrent No Change A05 Other bacterial foodborne intoxications, not elsewhere classified No Change Excludes1: Revise from Clostridium difficile foodborne intoxication and infection (A04.7) Revise to Clostridium difficile foodborne intoxication and infection (A04.7-) No Change Helminthiases (B65-B83) No Change B81 Other intestinal helminthiases, not elsewhere classified No Change Excludes1: Revise from angiostrongyliasis due to Parastrongylus cantonensis (B83.2) Revise to angiostrongyliasis due to: Add Angiostrongylus cantonensis (B83.2) Add Parastrongylus cantonensis (B83.2) No Change B81.3 Intestinal angiostrongyliasis Revise from Angiostrongyliasis due to Parastrongylus costaricensis Revise to Angiostrongyliasis due to: Add Angiostrongylus costaricensis Add Parastrongylus costaricensis No Change Chapter 2 No Change Neoplasms (C00-D49) No Change Malignant neoplasms of ill-defined, other secondary and unspecified sites (C76-C80) No Change C79 Secondary malignant neoplasm of other and unspecified sites Delete Excludes2: lymph node metastases (C77.0) No Change C79.1 Secondary malignant neoplasm of bladder -

Preventable Diseases Nick Wilson, Michael G Baker Abstract New Zealanders Can Now Reflect on and Celebrate 50 Years of Polio Elimination in This Country
THE NEW ZEALAND MEDICAL JOURNAL Journal of the New Zealand Medical Association Celebrating 50 years of polio elimination in New Zealand: but inadequate progress in eliminating other vaccine- preventable diseases Nick Wilson, Michael G Baker Abstract New Zealanders can now reflect on and celebrate 50 years of polio elimination in this country. This success was followed by eliminating two other infectious diseases, brucellosis and hydatids, and an imported potential disease vector, the southern saltmarsh mosquito. However, this country has made inadequate progress in eliminating several other vaccine-preventable diseases. These include measles, mumps, and rubella, which are priority candidates for elimination, and potentially Hib disease and rotavirus infection. To achieve such successes almost certainly requires that the country: (i) builds national leadership for elimination goals; (ii) develops detailed plans; (iii) continues recent successes in enhancing routine vaccination coverage; (iv) introduces rotavirus vaccine into the childhood immunisation schedule; and (v) strengthens surveillance and research (on such questions as the cost-effectiveness of new vaccines, measures to enhance uptake, and effective border controls to reduce the risk of disease importation). For 50 years now (since 1 April 1962), New Zealand has been free of transmission of wild-type polio virus infection. The end of this disease was particularly sudden with cases declining from 214 notifications in an outbreak in 1961 (with seven deaths) to only five cases in early 1962. 1 The end coincided with mass immunisation campaigns with the new Sabin (oral) vaccine in 1961 and 1962. The coverage in the vaccination campaign running from April to June 1962 was approximately 95% of the child population up to school leaving age. -

LIST of OCCUPATIONAL DISEASES (Revised 2010)
LIST OF OCCUPATIONAL DISEASES (revised 2010) Identification and recognition of occupational diseases: Criteria for incorporating diseases in the ILO list of occupational diseases Occupational Safety and Health Series, No. 74 List of occupational diseases (revised 2010) Identification and recognition of occupational diseases: Criteria for incorporating diseases in the ILO list of occupational diseases INTERNATIONAL LABOUR OFFICE • GENEVA Copyright © International Labour Organization 2010 First published 2010 Publications of the International Labour Office enjoy copyright under Protocol 2 of the Universal Copyright Convention. Nevertheless, short excerpts from them may be reproduced without authorization, on condition that the source is indicated. For rights of reproduction or translation, application should be made to ILO Publications (Rights and Permissions), International Labour Office, CH-1211 Geneva 22, Switzerland, or by email: pubdroit@ ilo.org. The International Labour Office welcomes such applications. Libraries, institutions and other users registered with reproduction rights organizations may make copies in accordance with the licences issued to them for this purpose. Visit www.ifrro.org to find the reproduction rights organization in your country. ILO List of occupational diseases (revised 2010). Identification and recognition of occupational diseases: Criteria for incorporating diseases in the ILO list of occupational diseases Geneva, International Labour Office, 2010 (Occupational Safety and Health Series, No. 74) occupational disease / definition. 13.04.3 ISBN 978-92-2-123795-2 ISSN 0078-3129 Also available in French: Liste des maladies professionnelles (révisée en 2010): Identification et reconnaissance des maladies professionnelles: critères pour incorporer des maladies dans la liste des maladies professionnelles de l’OIT (ISBN 978-92-2-223795-1, ISSN 0250-412x), Geneva, 2010, and in Spanish: Lista de enfermedades profesionales (revisada en 2010). -

ICD-10 International Statistical Classification of Diseases and Related Health Problems
ICD-10 International Statistical Classification of Diseases and Related Health Problems 10th Revision Volume 2 Instruction manual 2010 Edition WHO Library Cataloguing-in-Publication Data International statistical classification of diseases and related health problems. - 10th revision, edition 2010. 3 v. Contents: v. 1. Tabular list – v. 2. Instruction manual – v. 3. Alphabetical index. 1.Diseases - classification. 2.Classification. 3.Manuals. I.World Health Organization. II.ICD-10. ISBN 978 92 4 154834 2 (NLM classification: WB 15) © World Health Organization 2011 All rights reserved. Publications of the World Health Organization are available on the WHO web site (www.who.int) or can be purchased from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857; e-mail: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for noncommercial distribution – should be addressed to WHO Press through the WHO web site (http://www.who.int/about/licensing/copyright_form). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. -

Diseases of the Canine Digit
Diseases of the Canine Digit Diseases of the digit are relatively common and are particularly frustrating in terms of therapy. Unlike many other areas of skin, persisting diseases of the digit will almost always require biopsy to distinguish among a very long list of radically different etiologic possibilities. One cannot tell, just by looking at it, whether the digital swelling is chronic inflammation or squamous cell carcinoma, or whether the lump on the side of the digit is a harmless plasmacytoma or a potentially fatal amelanotic malignant melanoma. 1. Multiple nails that are brittle, deformed, or fall out: Most textbooks will provide a very long list of diseases of the nail and nail bed, but in practical terms I see only one syndrome: lupoid onychodystrophy. Bacterial and fungal paronychia, for example, is so rare in my collection that I have some skepticism that it even exists! The syndrome of lupoid onychodystrophy is seen in young mature dogs (1-5 years), and these animals present with a complaint of deformed nails that are periodically lost. The disease affects multiple digits on multiple feet, often eventually affecting all nails on all feet. The lesion is a lupus-like destructive disease (lymphocytic interface dermatitis with single cell necrosis) of the basal cells of the nail bed epithelium. As is true with similar histologic reactions affecting the nasal planum, gingiva, or conjunctiva, it is not yet clear whether this highly repeatable histologic pattern is really a reflection of a single disease, or is simply the way that the nail bed epithelium responds to a variety of different injuries. -

FAQ REGARDING DISEASE REPORTING in MONTANA | Rev
Disease Reporting in Montana: Frequently Asked Questions Title 50 Section 1-202 of the Montana Code Annotated (MCA) outlines the general powers and duties of the Montana Department of Public Health & Human Services (DPHHS). The three primary duties that serve as the foundation for disease reporting in Montana state that DPHHS shall: • Study conditions affecting the citizens of the state by making use of birth, death, and sickness records; • Make investigations, disseminate information, and make recommendations for control of diseases and improvement of public health to persons, groups, or the public; and • Adopt and enforce rules regarding the reporting and control of communicable diseases. In order to meet these obligations, DPHHS works closely with local health jurisdictions to collect and analyze disease reports. Although anyone may report a case of communicable disease, such reports are submitted primarily by health care providers and laboratories. The Administrative Rules of Montana (ARM), Title 37, Chapter 114, Communicable Disease Control, outline the rules for communicable disease control, including disease reporting. Communicable disease surveillance is defined as the ongoing collection, analysis, interpretation, and dissemination of disease data. Accurate and timely disease reporting is the foundation of an effective surveillance program, which is key to applying effective public health interventions to mitigate the impact of disease. What diseases are reportable? A list of reportable diseases is maintained in ARM 37.114.203. The list continues to evolve and is consistent with the Council of State and Territorial Epidemiologists (CSTE) list of Nationally Notifiable Diseases maintained by the Centers for Disease Control and Prevention (CDC). In addition to the named conditions on the list, any occurrence of a case/cases of communicable disease in the 20th edition of the Control of Communicable Diseases Manual with a frequency in excess of normal expectancy or any unusual incident of unexplained illness or death in a human or animal should be reported. -

ICD-10 International Statistical Classification of Diseases And
ICD-10 International statistical classification of diseases and related health problems 10th revision Volume 2 Instruction manual Fifth edition 2016 Volume 2.indb 1 11/09/15 10:46 WHO Library Cataloguing-in-Publication Data International statistical classification of diseases and related health problems. - 10th revision, Fifth edition, 2016. 3 v. Contents: v. 1. Tabular list -- v. 2. Instruction manual -- v. 3. Alphabetical index. 1.Diseases - classification. 2.Classification. 3.Manuals. I.World Health Organization. II.ICD-10. ISBN 978 92 4 154916 5 (NLM classification: WB 15) © World Health Organization 2011. Reprinted in 2015. All rights reserved. Publications of the World Health Organization are available on the WHO website (www.who.int) or can be purchased from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857; e-mail: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for non-commercial distribution – should be addressed to WHO Press through the WHO website (www.who.int/about/licensing/copyright_form/en/index.html). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted and dashed lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. -
Parkinson's Disease Occurs When a Group of Cells in the Area of the Brain Called the Substantia Nigra Begin to Malfunction and Die
Parkinson’s Disease Q &A sixth EDitiON Dear Friends: People affected by Parkinson’s disease — those living with Parkinson’s, their family members, their friends and the health care professionals that care for them — are all looking for answers to questions about the disease, its symptoms and treatments. This booklet is a compilation of the most frequently asked questions that the Parkinson’s Disease Foundation (PDF) receives through its National HelpLine. Some of these questions have simple answers, while others have complicated answers that are still evolving. While each question is answered as compre - hensively as possible, it is important to note that Parkinson’s is truly an individualized disease. Each person’s experience with Par kinson’s, including its symptoms and the rate at which it progresses, is different. Not all people living with Parkinson’s will experience all of the symptoms and side effects discussed in this booklet. Rather, each person may find that certain symptoms are more troublesome and may experience these symptoms at different points in the disease. If you have additional questions that you would like to discuss, please call PDF at ( 800) 457 - 6676 or email us at [email protected]. Sincerely, Robin Elliott President This publication was written and edited with the help of Parkinson’s disease specialists Blair Ford, M.D., of Columbia University Medical Center and W. Lawrence Severt, M.D., Ph.D., of Beth Israel Medical Center. table of contents I. Understanding Parkinson’s 5 – 31 • Overview • What Happens in Parkinson’s? • Motor Symptoms & Complications • Nonmotor Symptoms 2. T reating Parkinson’s 33 – 47 • Finding a Doctor • Medications & Surgical Treatments • Complementary & Alternative Therapies • Exercise & Nutrition 3. -

History of the Statistical Classification of Diseases and Causes of Death
Copyright information All material appearing in this report is in the public domain and may be reproduced or copied without permission; citation as to source, however, is appreciated. Suggested citation Moriyama IM, Loy RM, Robb-Smith AHT. History of the statistical classification of diseases and causes of death. Rosenberg HM, Hoyert DL, eds. Hyattsville, MD: National Center for Health Statistics. 2011. Library of Congress Cataloging-in-Publication Data Moriyama, Iwao M. (Iwao Milton), 1909-2006, author. History of the statistical classification of diseases and causes of death / by Iwao M. Moriyama, Ph.D., Ruth M. Loy, MBE, A.H.T. Robb-Smith, M.D. ; edited and updated by Harry M. Rosenberg, Ph.D., Donna L. Hoyert, Ph.D. p. ; cm. -- (DHHS publication ; no. (PHS) 2011-1125) “March 2011.” Includes bibliographical references. ISBN-13: 978-0-8406-0644-0 ISBN-10: 0-8406-0644-3 1. International statistical classification of diseases and related health problems. 10th revision. 2. International statistical classification of diseases and related health problems. 11th revision. 3. Nosology--History. 4. Death- -Causes--Classification--History. I. Loy, Ruth M., author. II. Robb-Smith, A. H. T. (Alastair Hamish Tearloch), author. III. Rosenberg, Harry M. (Harry Michael), editor. IV. Hoyert, Donna L., editor. V. National Center for Health Statistics (U.S.) VI. Title. VII. Series: DHHS publication ; no. (PHS) 2011- 1125. [DNLM: 1. International classification of diseases. 2. Disease-- classification. 3. International Classification of Diseases--history. 4. Cause of Death. 5. History, 20th Century. WB 15] RB115.M72 2011 616.07’8012--dc22 2010044437 For sale by the U.S. -

61St YEAR-61E ANNÉE
Wkly kpidem Rcc - Helete eputem. hebd. lÿM , 61, oü-76 No. 10 WORLD HEALTH ORGANIZATION ORGANISATION MONDIALE DE LA SANTÉ GENEVA GENÈVE WEEKLY EPIDEMIOLOGICAL RECORD RELEVÉ ÉPIDÉMIOLOGIQUE HEBDOMADAIRE Telegraphic Address, EP1DNATIONS GENEVA Telex m i l Adresse télégraphique: EP1DNAT10NS GENÈVE Télex 27S2I Automatic Telex Reply Service Service automatique de réponse par teks Telex 281SQ Geneva with ZC2SC u d ENGL for a reply ui English Télex 28150 Genève suivi de ZC2C et FRAN pour une réponse en français 7 MARCH 1986 61st YEAR-6 1 e ANNÉE 7 MARS 1986 ACQUIRED IMMUNODEFICIENCY SYNDROME SYNDROME DTMMUNODÉFICIT ACQUIS (AIDS) (SIDA) WHO/CDC case definition for AIDS Définition OMS/CDC du cas de SIDA For surveillance purposes, a relatively precise case definition is Aux fins de la surveillance, il est nécessaire de disposer d’une définition required that includes the most characteristic manifestations of relativement précise du cas de SIDA qui englobe les manifestations les LAV/HTLV-III infections. WHO recently adopted the case defi plus caractéristiques des infections par le LAV/HTLV-III. L’OMS a nition of AIDS in adults amd children developed by the Centers récemment adopté une définition du cas de SIDA chez l’adulte et l’enfant for Disease Control (CDC) and endorsed by the participants at the élaborée par les Centers for Disease Control (CDC) et entérinée par les Second Meeting of the WHO Collaborating Centres on AIDS held participants à la Deuxième réunion des Centres collaborateurs pour le in Geneva 16-18 December 1985. The WHO/CDC definition, to SIDA qui s’est tenue à Genève du 16au 18décembre 1985. -
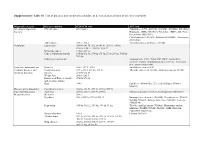
Supplementary Table S1. List of Diseases and Conditions Candidate to Be Tested As Predictors of One-Year Mortality
Supplementary Table S1. List of diseases and conditions candidate to be tested as predictors of one-year mortality Diagnostic category Disease/condition ICD-9 CM code ATC code Infectious and parasitic HIV infection 042.x-044.x Zidovudine (AZT) (J05AF01, J05AR01, J05AR04, J05AR05), diseases Didanosine (DDI) (J05AF02), Zalcitabine (DDC) (J05AF03), Pentamidine (P01CX01), Clarithromycin (J01FA09), Rifabutin (J04AB04), Atovaquone (P01AX06) Tuberculosis 010.x - 018.x Anti-tuberculosis antibiotics (J04AB) Neoplasms Lymphoma 200.00-202.38, 202.50-203.01, 203.8x, 238.6x, 273.3x, V10.71, V10.72, V10.79 Metastatic cancer 196.0x-199.1x Cancer, without metastasis 140.0x-172.9x, 174.0x-175.9x,179.x-195.8x, V10.0x- V10.9x Malignancy medication Antineoplastic (L01), Taxol (C07AB05), Interleukins (L03AC), Colony-stimulating factors (L03AA), Antinausea misc, ondansetron (A04) Endocrine, nutritional and Diabetes 250.x, 357.2, 362.0 Antidiabetic agents (A10) metabolic diseases, and Hypothyroidism 243.x-244.2, 244.8x, 244.9x Thyroid replacement (H03A), Antithyroid agents (H03B) immunity disorders Obesity 278.00-278.01 Weight loss 260.0x-263.9 Disorders of fluid, electrolyte, 276.0x-276.9x and acid-base balance Gout 274.x Colchicine (M04AC01), Uric acid inhibitors (M04AA, M04AB) Diseases of the blood and Coagulation defects 286.0x-286.9x, 287.1x, 287.3x-287.5x blood-forming organs Anaemias 280.0x, 280.1x-281.9x, 285.9x Marrow stimulants (L03AA), Erythropoietin (B03XA01) Mental disorders Dementia 290.x Psychosis 295.x-298.9x, 299.10-299.11 Butyrophenone derivates