Metabolic Checkpoints in Cancer Cell Cycle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DNA Damage Checkpoint Dynamics Drive Cell Cycle Phase Transitions
bioRxiv preprint doi: https://doi.org/10.1101/137307; this version posted August 4, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. DNA damage checkpoint dynamics drive cell cycle phase transitions Hui Xiao Chao1,2, Cere E. Poovey1, Ashley A. Privette1, Gavin D. Grant3,4, Hui Yan Chao1, Jeanette G. Cook3,4, and Jeremy E. Purvis1,2,4,† 1Department of Genetics 2Curriculum for Bioinformatics and Computational Biology 3Department of Biochemistry and Biophysics 4Lineberger Comprehensive Cancer Center University of North Carolina, Chapel Hill 120 Mason Farm Road Chapel Hill, NC 27599-7264 †Corresponding Author: Jeremy Purvis Genetic Medicine Building 5061, CB#7264 120 Mason Farm Road Chapel Hill, NC 27599-7264 [email protected] ABSTRACT DNA damage checkpoints are cellular mechanisms that protect the integrity of the genome during cell cycle progression. In response to genotoxic stress, these checkpoints halt cell cycle progression until the damage is repaired, allowing cells enough time to recover from damage before resuming normal proliferation. Here, we investigate the temporal dynamics of DNA damage checkpoints in individual proliferating cells by observing cell cycle phase transitions following acute DNA damage. We find that in gap phases (G1 and G2), DNA damage triggers an abrupt halt to cell cycle progression in which the duration of arrest correlates with the severity of damage. However, cells that have already progressed beyond a proposed “commitment point” within a given cell cycle phase readily transition to the next phase, revealing a relaxation of checkpoint stringency during later stages of certain cell cycle phases. -

Working on Genomic Stability: from the S-Phase to Mitosis
G C A T T A C G G C A T genes Review Working on Genomic Stability: From the S-Phase to Mitosis Sara Ovejero 1,2,3,* , Avelino Bueno 1,4 and María P. Sacristán 1,4,* 1 Instituto de Biología Molecular y Celular del Cáncer (IBMCC), Universidad de Salamanca-CSIC, Campus Miguel de Unamuno, 37007 Salamanca, Spain; [email protected] 2 Institute of Human Genetics, CNRS, University of Montpellier, 34000 Montpellier, France 3 Department of Biological Hematology, CHU Montpellier, 34295 Montpellier, France 4 Departamento de Microbiología y Genética, Universidad de Salamanca, Campus Miguel de Unamuno, 37007 Salamanca, Spain * Correspondence: [email protected] (S.O.); [email protected] (M.P.S.); Tel.: +34-923-294808 (M.P.S.) Received: 31 January 2020; Accepted: 18 February 2020; Published: 20 February 2020 Abstract: Fidelity in chromosome duplication and segregation is indispensable for maintaining genomic stability and the perpetuation of life. Challenges to genome integrity jeopardize cell survival and are at the root of different types of pathologies, such as cancer. The following three main sources of genomic instability exist: DNA damage, replicative stress, and chromosome segregation defects. In response to these challenges, eukaryotic cells have evolved control mechanisms, also known as checkpoint systems, which sense under-replicated or damaged DNA and activate specialized DNA repair machineries. Cells make use of these checkpoints throughout interphase to shield genome integrity before mitosis. Later on, when the cells enter into mitosis, the spindle assembly checkpoint (SAC) is activated and remains active until the chromosomes are properly attached to the spindle apparatus to ensure an equal segregation among daughter cells. -
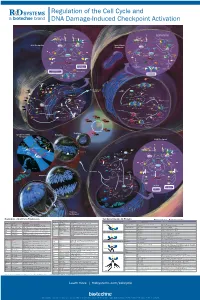
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Cell Cycle Checkpoint in Cancer: a Therapeutically Targetable Double-Edged Sword Roberta Visconti1*, Rosa Della Monica2,3 and Domenico Grieco2,3*
Visconti et al. Journal of Experimental & Clinical Cancer Research (2016) 35:153 DOI 10.1186/s13046-016-0433-9 REVIEW Open Access Cell cycle checkpoint in cancer: a therapeutically targetable double-edged sword Roberta Visconti1*, Rosa Della Monica2,3 and Domenico Grieco2,3* Abstract Major currently used anticancer therapeutics either directly damage DNA or target and upset basic cell division mechanisms like DNA replication and chromosome segregation. These insults elicit activation of cell cycle checkpoints, safeguard mechanisms that cells implement to correctly complete cell cycle phases, repair damage or eventually commit suicide in case damage is unrepairable. Although cancer cells appear to be advantageously defective in some aspects of checkpoint physiology, recent acquisitions on the biochemical mechanisms of the various checkpoints are offering new therapeutic approaches against cancer. Indeed, chemical manipulation of these mechanisms is providing new therapeutic strategies and tools to increase the killing efficacy of major cancer therapeutics as well as to directly promote cancer cell death. In this review we summarize developing concepts on how targeting cell cycle checkpoints may provide substantial improvement to cancer therapy. Keywords: Cell cycle checkpoint, Cancer drug, DNA damage, Mitosis, Chk1, Wee1, Spindle assembly checkpoint, Taxane, Vinca alkaloids, Fcp1 Background opportunities for cancer treatment. Here we will review The mechanisms of cell division and the genome itself how recent efforts to identify new strategies and drugs are routinely endangered by endogenous and exogenous targeting cell cycle checkpoints will likely translate soon insults. For instance reactive oxygen species, produced into benefit to clinical practice in oncology. As outlined during metabolic reactions, inflammation or exciting in Table 1, we will focus our attention on drugs targeting and ionizing radiations, can damage chromosomes and key players of the S and G2/M checkpoints activated in upset chromosome replication and segregation [1, 2]. -

Overexpression of CCDC69 Activates P14arf/MDM2/P53 Pathway and Confers Cisplatin Sensitivity Long Cui1,4* , Fang Zhou2, Cui Chen3 and Chi Chiu Wang4,5,6
Cui et al. Journal of Ovarian Research (2019) 12:4 https://doi.org/10.1186/s13048-019-0479-3 RESEARCH Open Access Overexpression of CCDC69 activates p14ARF/MDM2/p53 pathway and confers cisplatin sensitivity Long Cui1,4* , Fang Zhou2, Cui Chen3 and Chi Chiu Wang4,5,6 Abstract Objectives: The aim of the study is to explore the relationship between CCDC69 expression and resistance of ovarian cancer cells to cisplatin and reveal the underlying mechanism. Methods: One hundred thirty five ovarian cancer patients with intact chemo-response information from The Cancer Genome Atlas (TCGA) database were included and analyzed. Stable CCDC69 overexpressing 293 and ovarian cancer A2780 cell lines were established and subjected to examine cell apoptosis and cell cycle distribution using CCK-8 assay and flow cytometry. Cell cycle and apoptosis pathway were evaluated by immunoblots. Stability of p14ARF/MDM2/p53 pathway related proteins were determined by half-life analysis and ubiquitination experiments. Results: We found that CCDC69 expression was significantly higher in chemo-sensitive groups compared with chemo- resistant groups from TCGA database. High CCDC69 expression was associated longer survival. CCDC69 overexpressing 293 and A2780 cells with wildtype p53 and contributes to cisplatin sensitivity following treatment with cisplatin. We further found over-expression of CCDC69 activated p14ARF/MDM2/p53 pathway. Importantly, we also demonstrated that CCDC69 expression extended p53 and p14ARF protein half-life and shortened MDM2 protein half-life. Ubiquitination assay revealing a decrease in p14 ubiquitination in CCDC69 over-expression cells comparing to cells expressing empty vector. Conclusions: It is tempting to conclude that targeting CCDC69 may play a role in cisplatin resistance. -

The CDK1 Inhibitory Kinase MYT1 in DNA Damage Checkpoint Recovery
Oncogene (2013) 32, 4778–4788 & 2013 Macmillan Publishers Limited All rights reserved 0950-9232/13 www.nature.com/onc ORIGINAL ARTICLE The CDK1 inhibitory kinase MYT1 in DNA damage checkpoint recovery JPH Chow and RYC Poon Inhibition of cyclin-dependent kinase 1 (CDK1) by phosphorylation is a key regulatory mechanism for both the unperturbed cell cycle and the DNA damage checkpoint. Although both WEE1 and MYT1 can phosphorylate CDK1, little is known about the contribution of MYT1. We found that in contrast to WEE1, MYT1 was not important for the normal cell cycle or checkpoint activation. Time-lapse microscopy indicated that MYT1 did, however, have a rate-determining role during checkpoint recovery. Depletion of MYT1 induced precocious mitotic entry when the checkpoint was abrogated with inhibitors of either CHK1 or WEE1, indicating that MYT1 contributes to checkpoint recovery independently of WEE1. The acceleration of checkpoint recovery in MYT1-depleted cells was due to a lowering of threshold for CDK1 activation. The kinase activity of MYT1 was high during checkpoint activation and reduced during checkpoint recovery. Importantly, although depletion of MYT1 alone did not affect long- term cell growth, it potentiated with DNA damage to inhibit cell growth in clonogenic survival and tumor xenograft models. These results reveal the functions of MYT1 in checkpoint recovery and highlight the potential of MYT1 as a target for anti-cancer therapies. Oncogene (2013) 32, 4778–4788; doi:10.1038/onc.2012.504; published online 12 November 2012 Keywords: cell cycle; checkpoint; ionizing radiation INTRODUCTION CHK1/CHK2, which in turn inactivates CDC25 (reviewed in Chen 15 Cyclin-dependent kinase 1 (CDK1) is one of the major kinases for and Poon ). -

Phosphatases Reverse P53-Mediated Cell Cycle Checkpoints
COMMENTARY Phosphatases reverse p53-mediated cell cycle checkpoints Lawrence A. Donehower1 for the damage or dysfunction to be repaired Departments of Molecular Virology and Microbiology, Molecular, and Cellular Biology, Pediatrics without being transmitted to progeny cells. and the Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030 Thus, p53 has been called the “guardian of the genome” (3).Giventheimportanceof p53 as a cellular failsafe mechanism, it is The Cancer Genome Atlas and other large- In the normal cell, the p53 protein func- not surprising that its inactivation is a highly scale cancer genome sequencing projects tions as a stress integrator that becomes selected event in cancer progression. have identified an impressive number of activated in response to myriad dysfunctional Activated p53 can halt cell division in both states (e.g., damaged DNA, aberrant onco- significantly mutated genes that are likely the G1 and G2 phases of the cell division genic signaling, and hypoxia). Once activated, drivers of cancer progression (1). These stud- cycle. G1 is the preparation phase of the cell p53 can initiate stress response programs to ies have definitively confirmed that the gene before replication of its DNA and G2 pre- eliminate the dysfunctional state, such as encoding the p53 tumor suppressor is the pares the cell for mitosis. Arrest and repair of repair of DNA damage. If the stressed cell most frequently mutated gene in human can- cells before DNA replication or mitosis are is dividing, p53 can enact any one of several cers. A major component of the p53 antican- likely to prevent damage-induced mutational antiproliferative programs. -

Cell Cycle Arrest Through Indirect Transcriptional Repression by P53: I Have a DREAM
Cell Death and Differentiation (2018) 25, 114–132 Official journal of the Cell Death Differentiation Association OPEN www.nature.com/cdd Review Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM Kurt Engeland1 Activation of the p53 tumor suppressor can lead to cell cycle arrest. The key mechanism of p53-mediated arrest is transcriptional downregulation of many cell cycle genes. In recent years it has become evident that p53-dependent repression is controlled by the p53–p21–DREAM–E2F/CHR pathway (p53–DREAM pathway). DREAM is a transcriptional repressor that binds to E2F or CHR promoter sites. Gene regulation and deregulation by DREAM shares many mechanistic characteristics with the retinoblastoma pRB tumor suppressor that acts through E2F elements. However, because of its binding to E2F and CHR elements, DREAM regulates a larger set of target genes leading to regulatory functions distinct from pRB/E2F. The p53–DREAM pathway controls more than 250 mostly cell cycle-associated genes. The functional spectrum of these pathway targets spans from the G1 phase to the end of mitosis. Consequently, through downregulating the expression of gene products which are essential for progression through the cell cycle, the p53–DREAM pathway participates in the control of all checkpoints from DNA synthesis to cytokinesis including G1/S, G2/M and spindle assembly checkpoints. Therefore, defects in the p53–DREAM pathway contribute to a general loss of checkpoint control. Furthermore, deregulation of DREAM target genes promotes chromosomal instability and aneuploidy of cancer cells. Also, DREAM regulation is abrogated by the human papilloma virus HPV E7 protein linking the p53–DREAM pathway to carcinogenesis by HPV.Another feature of the pathway is that it downregulates many genes involved in DNA repair and telomere maintenance as well as Fanconi anemia. -

A Haploid Genetic Screen Identifies the G1/S Regulatory Machinery As a Determinant of Wee1 Inhibitor Sensitivity
A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity Anne Margriet Heijinka, Vincent A. Blomenb, Xavier Bisteauc, Fabian Degenera, Felipe Yu Matsushitaa, Philipp Kaldisc,d, Floris Foijere, and Marcel A. T. M. van Vugta,1 aDepartment of Medical Oncology, University Medical Center Groningen, University of Groningen, 9723 GZ Groningen, The Netherlands; bDivision of Biochemistry, The Netherlands Cancer Institute, 1066 CX Amsterdam, The Netherlands; cInstitute of Molecular and Cell Biology, Agency for Science, Technology and Research, Proteos#3-09, Singapore 138673, Republic of Singapore; dDepartment of Biochemistry, National University of Singapore, Singapore 117597, Republic of Singapore; and eEuropean Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, 9713 AV Groningen, The Netherlands Edited by Stephen J. Elledge, Harvard Medical School, Boston, MA, and approved October 21, 2015 (received for review March 17, 2015) The Wee1 cell cycle checkpoint kinase prevents premature mitotic Wee1 kinase at tyrosine (Tyr)-15 to prevent unscheduled Cdk1 entry by inhibiting cyclin-dependent kinases. Chemical inhibitors activity (5, 6). Conversely, timely activation of Cdk1 depends on of Wee1 are currently being tested clinically as targeted anticancer Tyr-15 dephosphorylation by one of the Cdc25 phosphatases drugs. Wee1 inhibition is thought to be preferentially cytotoxic in (7–10). When DNA is damaged, the downstream DNA damage p53-defective cancer cells. However, TP53 mutant cancers do not response (DDR) kinases Chk1 and Chk2 inhibit Cdc25 phos- respond consistently to Wee1 inhibitor treatment, indicating the phatases through direct phosphorylation, which blocks Cdk1 existence of genetic determinants of Wee1 inhibitor sensitivity other activation (11–13). -

Lnvited Review Gain of Function Properties of Mutant P53 Proteins at the Mitotic Spindle Cell Cycle Checkpoint
HistolHistopathol (2000) 15: 551-556 Histology and 001: 10.14670/HH-15.551 Histopathology http://www.hh.um .es Ce/fular and Molecular Biology lnvited Review Gain of function properties of mutant p53 proteins at the mitotic spindle cell cycle checkpoint M.L. Hixon, A. Flores, M. Wagner and A. Gualberto Departmentá lbysiologyand Biophysics,Case WesternReserve UniversitySchool of Medicine,Cleveland, Ohio, IB<\. Summary. Mutations in the p53 tumor suppressor gene malignant phenotype. Therefore, it is becoming locus predispose human cells to chromosomal instability. increasingly evident that loss of the mechanisms that This is due in part to interference of mutant p53 proteins maintain the integrity of the human genome is a primary with the activity of the mitotic spindle and postmitotic event in the genesis of cancer. cell cycle checkpoints. Recent data demonstrates that Research addressing the relationship between cell wild type p53 is required far postmitotic checkpoint proliferation and the maintenance of genomic integrity activity, but plays no role at the mitotic spindle in human cells is relatively recent. Extensive evidence checkpoint. Likewise, structural dominant p53 mutants demonstrates that the onset of every phase of the cell demonstrate gain-of-function properties at the mitotic cycle depends critically on the adequate completion of spindle checkpoint and dominant negative properties at the previous cell cycle phase. The coordination of these the postmitotic checkpoint. At mitosis, mutant p53 events is carried out by pathways called cell cycle proteins interfere with the control of the metaphase-to checkpoints (Elledge, 1996). These pathways oversee anaphase progression by up-regulating the expression of the progression of the cell cycle and play a crucial role CKsl, a protein that mediates activatory phosphorylation in the maintenance of genomic integrity by coordinating of the anaphase promoting complex (APC) by Cdc2. -

Recent Advances of Cell-Cycle Inhibitor Therapies for Pediatric Cancer Christopher C
Published OnlineFirst November 2, 2017; DOI: 10.1158/0008-5472.CAN-17-2066 Cancer Review Research Recent Advances of Cell-Cycle Inhibitor Therapies for Pediatric Cancer Christopher C. Mills1, EA. Kolb2, and Valerie B. Sampson2 Abstract This review describes the pivotal roles of cell-cycle and check- MLN8237), Wee1 kinase (MK-1775), KSP (ispinesib), and tubu- point regulators and discusses development of specific cell-cycle lin (taxanes, vinca alkaloids), are presented. The design of mech- inhibitors for therapeutic use for pediatric cancer. The mechanism anism-based combinations that exploit the cross-talk of signals of action as well as the safety and tolerability of drugs in pediatric activated by cell-cycle arrest, as well as pediatric-focused drug patients, including compounds that target CDK4/CDK6 (palbo- development, are critical for the advancement of drugs for rare ciclib, ribociclib, and abemaciclib), aurora kinases (AT9283 and childhood diseases. Cancer Res; 77(23); 6489–98. Ó2017 AACR. Introduction of DNA damage, epigenetics, metabolism, proteolytic degra- dation, stem cell self-renewal, neuronal functions, and sper- Recent preclinical and clinical studies of highly selective matogenesis (2). agents that target various regulators of the mammalian cell Selective members of the CDK family of protein kinases act as cycle demonstrate cell-cycle arrest, inhibition of transcription, oncogenic stimuli in several types of cancer (e.g., CDK1 in breast and apoptotic cell death in models of human cancer. The cell- cancer and colon cancer (3, 4), CDK4 in familial melanoma (5), cycle drives proliferation of cells by the duplication of chromo- and CDK6 in MLL-rearranged leukemia (6). -

The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer
International Journal of Molecular Sciences Review The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer 1,2, 1,2, 1,2, 1,2 1,2 Lei Ding y, Jiaqi Cao y, Wen Lin y, Hongjian Chen , Xianhui Xiong , Hongshun Ao 1,2, Min Yu 1,2, Jie Lin 1,2 and Qinghua Cui 1,2,* 1 Lab of Biochemistry & Molecular Biology, School of Life Sciences, Yunnan University, Kunming 650091, China; [email protected] (L.D.); [email protected] (J.C.); [email protected] (W.L.); [email protected] (H.C.); [email protected] (X.X.); [email protected] (H.A.); [email protected] (M.Y.); [email protected] (J.L.) 2 Key Lab of Molecular Cancer Biology, Yunnan Education Department, Kunming 650091, China * Correspondence: [email protected] The authors contributed equally to this work. y Received: 31 December 2019; Accepted: 24 February 2020; Published: 13 March 2020 Abstract: Cyclin-dependent kinases (CDKs) are serine/threonine kinases whose catalytic activities are regulated by interactions with cyclins and CDK inhibitors (CKIs). CDKs are key regulatory enzymes involved in cell proliferation through regulating cell-cycle checkpoints and transcriptional events in response to extracellular and intracellular signals. Not surprisingly, the dysregulation of CDKs is a hallmark of cancers, and inhibition of specific members is considered an attractive target in cancer therapy. In breast cancer (BC), dual CDK4/6 inhibitors, palbociclib, ribociclib, and abemaciclib, combined with other agents, were approved by the Food and Drug Administration (FDA) recently for the treatment of hormone receptor positive (HR+) advanced or metastatic breast cancer (A/MBC), as well as other sub-types of breast cancer.