Tyrosine Phosphorylation: from Discovery to the Kinome and Beyond
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PERK Antibody / EIF2AK3 (RQ4206)
PERK Antibody / EIF2AK3 (RQ4206) Catalog No. Formulation Size RQ4206 0.5mg/ml if reconstituted with 0.2ml sterile DI water 100 ug Bulk quote request Availability 1-3 business days Species Reactivity Human, Mouse, Rat Format Antigen affinity purified Clonality Polyclonal (rabbit origin) Isotype Rabbit IgG Purity Antigen affinity purified Buffer Lyophilized from 1X PBS with 2% Trehalose and 0.025% sodium azide UniProt Q9NZJ5 Applications Western Blot : 0.5-1ug/ml Flow cytometry : 1-3ug/10^6 cells Direct ELISA : 0.1-0.5ug/ml Limitations This PERK antibody is available for research use only. Western blot testing of human 1) HeLa, 2) COLO320, 3) A549, 4) SK-OV-3, 5) A431, 6) rat brain and 7) mouse brain lysate with PERK antibody at 0.5ug/ml. Predicted molecular weight ~125 kDa, observed here at ~140 kDa. Flow cytometry testing of human HepG2 cells with PERK antibody at 1ug/10^6 cells (blocked with goat sera); Red=cells alone, Green=isotype control, Blue= PERK antibody. Description Eukaryotic translation initiation factor 2-alpha kinase 3, also known as protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), is an enzyme that in humans is encoded by the EIF2AK3 gene. The protein encoded by this gene phosphorylates the alpha subunit of eukaryotic translation-initiation factor 2, leading to its inactivation, and thus to a rapid reduction of translational initiation and repression of global protein synthesis. This protein is thought to modulate mitochondrial function. It is a type I membrane protein located in the endoplasmic reticulum (ER), where it is induced by ER stress caused by malfolded proteins. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

De Novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation
bioRxiv preprint doi: https://doi.org/10.1101/757039; this version posted September 16, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. De novo EIF2AK1 and EIF2AK2 variants are associated with developmental delay, leukoencephalopathy, and neurologic decompensation Dongxue Mao1,2, Chloe M. Reuter3,4, Maura R.Z. Ruzhnikov5,6, Anita E. Beck7, Emily G. Farrow8,9,10, Lisa T. Emrick1,11,12,13, Jill A. Rosenfeld12, Katherine M. Mackenzie5, Laurie Robak2,12,13, Matthew T. Wheeler3,14, Lindsay C. Burrage12,13, Mahim Jain15, Pengfei Liu12, Daniel Calame11,13, Sebastien Küry17,18, Martin Sillesen19, Klaus Schmitz-Abe20, Davide Tonduti21, Luigina Spaccini22, Maria Iascone23, Casie A. Genetti20, Madeline Graf16, Alyssa Tran12, Mercedes Alejandro12, Undiagnosed Diseases Network, Brendan H. Lee12,13, Isabelle Thiffault8,9,24, Pankaj B. Agrawal#,20, Jonathan A. Bernstein#,3,25, Hugo J. Bellen#,2,12,26,27,28, Hsiao- Tuan Chao#,1,2,11,12,13,28,27,29 #Correspondence should be addressed: [email protected] (P.A.), [email protected] (J.A.B.), [email protected] (H.J.B.), [email protected] (H.T.C.) 1Department of Pediatrics, Baylor College of Medicine (BCM), Houston, TX 2Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 3Stanford Center for Undiagnosed Diseases, Stanford University, Stanford, CA 4Stanford Center for Inherited Cardiovascular Disease, Division of Cardiovascular Medicine, -

Coordinate Phosphorylation of Multiple Residues on Single AKT1 and AKT2 Molecules
Oncogene (2014) 33, 3463–3472 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc ORIGINAL ARTICLE Coordinate phosphorylation of multiple residues on single AKT1 and AKT2 molecules H Guo1,6, M Gao1,6,YLu1, J Liang1, PL Lorenzi2, S Bai3, DH Hawke4,JLi1, T Dogruluk5, KL Scott5, E Jonasch3, GB Mills1 and Z Ding1 Aberrant AKT activation is prevalent across multiple human cancer lineages providing an important new target for therapy. Twenty- two independent phosphorylation sites have been identified on specific AKT isoforms likely contributing to differential isoform regulation. However, the mechanisms regulating phosphorylation of individual AKT isoform molecules have not been elucidated because of the lack of robust approaches able to assess phosphorylation of multiple sites on a single AKT molecule. Using a nanofluidic proteomic immunoassay (NIA), consisting of isoelectric focusing followed by sensitive chemiluminescence detection, we demonstrate that under basal and ligand-induced conditions that the pattern of phosphorylation events is markedly different between AKT1 and AKT2. Indeed, there are at least 12 AKT1 peaks and at least 5 AKT2 peaks consistent with complex combinations of phosphorylation of different sites on individual AKT molecules. Following insulin stimulation, AKT1 was phosphorylated at Thr308 in the T-loop and Ser473 in the hydrophobic domain. In contrast, AKT2 was only phosphorylated at the equivalent sites (Thr309 and Ser474) at low levels. Further, Thr308 and Ser473 phosphorylation occurred predominantly on the same AKT1 molecules, whereas Thr309 and Ser474 were phosphorylated primarily on different AKT2 molecules. Although basal AKT2 phosphorylation was sensitive to inhibition of phosphatidylinositol 3-kinase (PI3K), basal AKT1 phosphorylation was essentially resistant. -
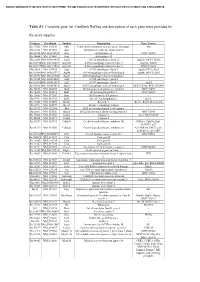
Table S1. Complete Gene List. Genbank Refseq and Description of Each Gene Were Provided By
Document downloaded from http://www.elsevier.es, day 24/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Table S1. Complete gene list. GenBank RefSeq and description of each gene were provided by the array supplier. Unigene GeneBank Symbol Description Gene Name/s Rn.11422 NM_033230 Akt1 V-akt murine thymoma viral oncogene homolog 1 Akt Rn.2104 NM_019288 App Amyloid beta (A4) precursor protein - Rn.23323 NM_001034933 Arsa Arylsulfatase A MGC125207 Rn.94004 NM_033443 Arsb Arylsulfatase B - Rn.6224 NM_001038495 Atg12 ATG12 autophagy related 12 Apg12l, MGC125080 Rn.101734NM_001108809 Atg16l1 ATG16 autophagy related 16-like 1 Apg16l, Wdr30 Rn.104199NM_001191560 Atg16l2 ATG16 autophagy related 16-like 2 RGD1311400 Rn.3084 NM_134394 Atg3 ATG3 autophagy related 3 Apg3l, PIG-1, Pig1 Rn.163086NM_001025711 Atg4b ATG4 autophagy related 4 homolog B Apg4b, MGC112887 Rn.23378 NM_001107948 Atg4c ATG4 autophagy related 4 homolog C - Rn.98385 NM_001014250 Atg5 ATG5 autophagy related 5 - Rn.162765NM_001012097 Atg7 ATG7 autophagy related 7 Apg7l Rn.35248 NM_001014218 Atg9a ATG9 autophagy related 9 homolog A MGC105908, RGD1310450 Rn.36696 NM_022698 Bad BCL2-associated agonist of cell death MGC72439 Rn.14598 NM_053812 Bak1 BCL2-antagonist/killer 1 MGC108627 Rn.10668 NM_017059 Bax Bcl2-associated X protein - Rn.9996 NM_016993 Bcl2 B-cell CLL/lymphoma 2 Bcl-2 Rn.10323 NM_031535 Bcl2l1 Bcl2-like 1 Bcl-xl, Bcl2l, Bclx, bcl-X Rn.2776 NM_053739 Becn1 Beclin 1, autophagy related - Rn.31142 NM_022684 -

Divergent Regulation of Akt1 and Akt2 Isoforms in Insulin Target Tissues of Obese Zucker Rats Young-Bum Kim, Odile D
Divergent Regulation of Akt1 and Akt2 Isoforms in Insulin Target Tissues of Obese Zucker Rats Young-Bum Kim, Odile D. Peroni, Thomas F. Franke, and Barbara B. Kahn To determine whether impaired Akt (protein kinase B or rac) activation contributes to insulin resistance in vivo, we examined the expression, phosphorylation, he effect of insulin to acutely stimulate glucose and kinase activities of Akt1 and Akt2 isoforms in uptake and metabolism in peripheral tissues is insulin target tissues of insulin-resistant obese Zucker essential for normal glucose homeostasis. Resis- ؋ rats. In lean rats, insulin (10 U/kg i.v. 2.5 min) stim- tance to this effect is a major pathogenic feature ulated Akt1 activity 6.2-, 8.8-, and 4.4-fold and Akt2 T of type 2 diabetes (1,2) and contributes to the morbidity of activity 5.4-, 9.3-, and 1.8-fold in muscle, liver, and adi- pose tissue, respectively. In obese rats, insulin-stimu- obesity as well as type 1 diabetes (3). Whereas many of the lated Akt1 activity decreased 30% in muscle and 21% proximal steps in insulin signaling have been identified, the in adipose tissue but increased 37% in liver compared downstream pathways for insulin action to maintain glucose with lean littermates. Insulin-stimulated Akt2 activity homeostasis are still unknown. Insulin action involves a decreased 29% in muscle and 37% in liver but series of signaling cascades initiated by insulin binding to its increased 24% in adipose tissue. Akt2 protein levels receptor and eliciting receptor autophosphorylation and acti- were reduced 56% in muscle and 35% in liver of obese vation of receptor tyrosine kinases, which result in tyrosine rats, but Akt1 expression was unaltered. -

LIM and Cysteine-Rich Domains 1 (LMCD1) Regulates Skeletal Muscle Hypertrophy, Calcium Handling, and Force Duarte M
Ferreira et al. Skeletal Muscle (2019) 9:26 https://doi.org/10.1186/s13395-019-0214-1 RESEARCH Open Access LIM and cysteine-rich domains 1 (LMCD1) regulates skeletal muscle hypertrophy, calcium handling, and force Duarte M. S. Ferreira1, Arthur J. Cheng2,3, Leandro Z. Agudelo1,4, Igor Cervenka1, Thomas Chaillou2,5, Jorge C. Correia1, Margareta Porsmyr-Palmertz1, Manizheh Izadi1,6, Alicia Hansson1, Vicente Martínez-Redondo1, Paula Valente-Silva1, Amanda T. Pettersson-Klein1, Jennifer L. Estall7, Matthew M. Robinson8, K. Sreekumaran Nair8, Johanna T. Lanner2 and Jorge L. Ruas1* Abstract Background: Skeletal muscle mass and strength are crucial determinants of health. Muscle mass loss is associated with weakness, fatigue, and insulin resistance. In fact, it is predicted that controlling muscle atrophy can reduce morbidity and mortality associated with diseases such as cancer cachexia and sarcopenia. Methods: We analyzed gene expression data from muscle of mice or human patients with diverse muscle pathologies and identified LMCD1 as a gene strongly associated with skeletal muscle function. We transiently expressed or silenced LMCD1 in mouse gastrocnemius muscle or in mouse primary muscle cells and determined muscle/cell size, targeted gene expression, kinase activity with kinase arrays, protein immunoblotting, and protein synthesis levels. To evaluate force, calcium handling, and fatigue, we transduced the flexor digitorum brevis muscle with a LMCD1-expressing adenovirus and measured specific force and sarcoplasmic reticulum Ca2+ release in individual fibers. Finally, to explore the relationship between LMCD1 and calcineurin, we ectopically expressed Lmcd1 in the gastrocnemius muscle and treated those mice with cyclosporine A (calcineurin inhibitor). In addition, we used a luciferase reporter construct containing the myoregulin gene promoter to confirm the role of a LMCD1- calcineurin-myoregulin axis in skeletal muscle mass control and calcium handling. -

In Vitro and in Vivo Profile of a Selective, Allosteric Akt2 Inhibitor
In Vitro and In Vivo Profile of a Selective, Allosteric Akt2 Inhibitor Martin O'Rourke1, Caroline Boyd1, Estelle McLean1, Natalie Page1, Shane Rountree1, Steven Shepherd1, Mary McFarland1 Colin O’Dowd1, Graham Trevitt1, Iain James1, Timothy Harrison1,2, Ultan McDermott3 1Almac Discovery, Craigavon, United Kingdom; 2Centre for Cancer Research and Cell Biology, Queens University, Belfast, United Kingdom 3 Wellcome Trust Sanger Institute, Hinxton, Cambs, CB10 1SA Biochemical efficacy of ADC-0008830, an Akt2 selective Pathway biomarker Overview inhibitor engagement in vitro Figure 1: Dose dependent inhibition of a) Akt1 b) Akt2 and c) Akt3 in an FP Figure 2: Inhibition of pathway • Akt protein family members (Akt biochemical assay biomarkers by ELISA 1-3) are involved in a wide variety a b c pGSK3β pPRAS 40 of biological processes including Cell line pAkt (nM) cell proliferation, apoptosis, and (nM) (nM) tumorigenesis. PC3 60 100 15 (IC50 7,600nM) (IC 4,200nM) (IC 27nM) • We have identified a novel, 50 50 LnCaP 500 6000 3600 allosteric Akt 2 selective inhibitor, A2780 160 740 840 which exhibits single agent A549 130 129 150 efficacy in a panel of cell-lines. Panc-1 9 234 Not tested • In collaboration with the Sanger • ADC-0008830 is a potent inhibitor of the Akt2 isoform Institute, we have identified genes • ADC-0008830 inhibits key • No activity observed against PH domain null Akt isoforms confirming of sensitivity and resistance for components of the Akt pathway an allosteric mode of action subtype selective Akt inhibitors in multiple cell lines • Excellent selectivity against a kinase counterscreen panel (data not shown) Dose and time dependent inhibition of Cellular profiling of ADC-0008939, a prototypical isoform selective Akt2 pathway biomarkers (Panc-1) inhibitor, in the Sanger cell line panel Figure 3: Dose and time dependent inhibition of Akt Figure 4: Pharmacogenomic analysis in 432 cell lines of ADC-0008939, a prototypical isoform selective pathway in Panc-1 cell line Akt inhibitor (IC50 Akt1 300nM; Akt2 7nM; Akt3 1230nM). -

Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma
G C A T T A C G G C A T genes Article Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma Dumitru A. Iacobas Personalized Genomics Laboratory, CRI Center for Computational Systems Biology, Roy G Perry College of Engineering, Prairie View A&M University, Prairie View, TX 77446, USA; [email protected]; Tel.: +1-936-261-9926 Received: 1 August 2020; Accepted: 1 September 2020; Published: 2 September 2020 Abstract: Publicly available (own) transcriptomic data have been analyzed to quantify the alteration in functional pathways in thyroid cancer, establish the gene hierarchy, identify potential gene targets and predict the effects of their manipulation. The expression data have been generated by profiling one case of papillary thyroid carcinoma (PTC) and genetically manipulated BCPAP (papillary) and 8505C (anaplastic) human thyroid cancer cell lines. The study used the genomic fabric paradigm that considers the transcriptome as a multi-dimensional mathematical object based on the three independent characteristics that can be derived for each gene from the expression data. We found remarkable remodeling of the thyroid hormone synthesis, cell cycle, oxidative phosphorylation and apoptosis pathways. Serine peptidase inhibitor, Kunitz type, 2 (SPINT2) was identified as the Gene Master Regulator of the investigated PTC. The substantial increase in the expression synergism of SPINT2 with apoptosis genes in the cancer nodule with respect to the surrounding normal tissue (NOR) suggests that SPINT2 experimental overexpression may force the PTC cells into apoptosis with a negligible effect on the NOR cells. The predictive value of the expression coordination for the expression regulation was validated with data from 8505C and BCPAP cell lines before and after lentiviral transfection with DDX19B. -

Supplemental Table 1: Snps Genotyped for NCO, Listed Alphabetically by Gene Name
Supplemental Table 1: SNPs genotyped for NCO, listed alphabetically by gene name. Gene Name SNP rs# ACVR1 rs10497189 ACVR1 rs10497191 ACVR1 rs10497192 ACVR1 rs10933441 ACVR1 rs1146035 ACVR1 rs1220110 ACVR1 rs17182166 ACVR1 rs17798043 ACVR1 rs4380178 ACVR1 rs6719924 ACVR2 rs1128919 ACVR2 rs10497025 ACVR2 rs1424941 ACVR2 rs1424954 ACVR2 rs17742573 ACVR2 rs2382112 ACVR2 rs4419186 AKT2 rs11671439 AKT2 rs12460555 AKT2 rs16974157 AKT2 rs2304188 AKT2 rs3730050 AKT2 rs7250897 AKT2 rs7254617 AKT2 rs874269 ALOX12B/ALOXE3 rs3809881 ALOX15B rs4792147 ALOX15B rs9898751 AMH rs10407022 AMH rs2074860 AMH rs3746158 AMH rs4806834 AMH rs757595 AMH rs886363 AMHR2 rs10876451 AMHR2 rs11170547 AMHR2 rs11170558 APC rs2229992 APC rs351771 APC rs41115 APC rs42427 APC rs459552 APC rs465899 APEX1 rs2275007 APEX1 rs3136820 15 APEX1 rs938883 APEX1 rs11160711 APEX1 rs1713459 APEX1 rs1713460 APEX1 rs1760941 APEX1 rs2275008 APEX1 rs3120073 APEX1 rs4465523 AR rs12011793 AR rs1204038 AR rs1337080 AR rs1337082 AR rs2207040 AR rs2361634 AR rs5031002 AR rs6152 AR rs962458 ATM rs1800058 ATM rs1800889 ATM rs11212570 ATM rs17503908 ATM rs227060 ATM rs228606 ATM rs3092991 ATM rs4987876 ATM rs4987886 ATM rs4987923 ATM rs4988023 ATM rs611646 ATM rs639923 ATM rs672655 ATR rs2229032 ATR rs10804682 ATR rs11920625 ATR rs13065800 ATR rs13085998 ATR rs13091637 ATR rs1802904 ATR rs6805118 ATR rs9856772 BACH1 rs388707 BACH1 rs1153276 BACH1 rs1153280 BACH1 rs1153284 BACH1 rs1153285 BACH1 rs17743655 BACH1 rs17744121 BACH1 rs2300301 BACH1 rs2832283 16 BACH1 rs411697 BACH1 rs425989 BARD1 rs1048108 -

Activation of Diverse Signalling Pathways by Oncogenic PIK3CA Mutations
ARTICLE Received 14 Feb 2014 | Accepted 12 Aug 2014 | Published 23 Sep 2014 DOI: 10.1038/ncomms5961 Activation of diverse signalling pathways by oncogenic PIK3CA mutations Xinyan Wu1, Santosh Renuse2,3, Nandini A. Sahasrabuddhe2,4, Muhammad Saddiq Zahari1, Raghothama Chaerkady1, Min-Sik Kim1, Raja S. Nirujogi2, Morassa Mohseni1, Praveen Kumar2,4, Rajesh Raju2, Jun Zhong1, Jian Yang5, Johnathan Neiswinger6, Jun-Seop Jeong6, Robert Newman6, Maureen A. Powers7, Babu Lal Somani2, Edward Gabrielson8, Saraswati Sukumar9, Vered Stearns9, Jiang Qian10, Heng Zhu6, Bert Vogelstein5, Ben Ho Park9 & Akhilesh Pandey1,8,9 The PIK3CA gene is frequently mutated in human cancers. Here we carry out a SILAC-based quantitative phosphoproteomic analysis using isogenic knockin cell lines containing ‘driver’ oncogenic mutations of PIK3CA to dissect the signalling mechanisms responsible for oncogenic phenotypes induced by mutant PIK3CA. From 8,075 unique phosphopeptides identified, we observe that aberrant activation of PI3K pathway leads to increased phosphorylation of a surprisingly wide variety of kinases and downstream signalling networks. Here, by integrating phosphoproteomic data with human protein microarray-based AKT1 kinase assays, we discover and validate six novel AKT1 substrates, including cortactin. Through mutagenesis studies, we demonstrate that phosphorylation of cortactin by AKT1 is important for mutant PI3K-enhanced cell migration and invasion. Our study describes a quantitative and global approach for identifying mutation-specific signalling events and for discovering novel signalling molecules as readouts of pathway activation or potential therapeutic targets. 1 McKusick-Nathans Institute of Genetic Medicine and Department of Biological Chemistry, Johns Hopkins University School of Medicine, 733 North Broadway, BRB 527, Baltimore, Maryland 21205, USA. -

Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action
International Journal of Molecular Sciences Review Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action Paweł Łukasik 1, Irena Baranowska-Bosiacka 2, Katarzyna Kulczycka 3 and Izabela Gutowska 1,* 1 Department of Medical Chemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] 2 Department of Biochemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] 3 Department of Pharmaceutical Chemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] * Correspondence: [email protected] Abstract: Recent studies on cyclin-dependent kinase (CDK) inhibitors have revealed that small molecule drugs have become very attractive for the treatment of cancer and neurodegenerative disorders. Most CDK inhibitors have been developed to target the ATP binding pocket. However, CDK kinases possess a very similar catalytic domain and three-dimensional structure. These features make it difficult to achieve required selectivity. Therefore, inhibitors which bind outside the ATP binding site present a great interest in the biomedical field, both from the fundamental point of view and for the wide range of their potential applications. This review tries to explain whether the ATP competitive inhibitors are still an option for future research, and highlights alternative approaches to discover more selective and potent small molecule inhibitors. Keywords: cyclin-dependent kinase inhibitors; cancer; cell cycle; CDKs; CDK inhibitors Citation: Łukasik, P.; Baranowska-Bosiacka, I.; Kulczycka, K.; Gutowska, I. Inhibitors of Cyclin-Dependent Kinases: Types 1. Cyclin-Dependent Kinases (CDKs) and Their Mechanism of Action.