Accepted Manuscript
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Thysanoptera: Phlaeothripidae): Redefinition and Key to Species
The southern Palaearctic genus Neoheegeria (Thysanoptera: Phlaeothripidae): redefinition and key to species Kambiz Minaei, Parvaneh Azemayeshfard & Laurence A. Mound Problems in character state definition and interpretation in the Haplothrips-group are discussed, together with their implications for species identification and systematics. As a result, Neoheegeria Schmutz, 1909 is redefined to include only those species in this group that have three sensoria on the third antennal segment. The subgenus Haplothrips (Gigaplothrips) Priesner, 1949 is synonymised with Neoheegeria, and four species are recognized as valid; N. dalmatica Schmutz, 1909, N. gigantea (Priesner, 1934) comb.n., N. persica Priesner, 1954, and N. sinaitica Priesner, 1934. Three new synonyms are recognized under N. dalmatica; N. ballotae Priesner, 1951, N. hamanni Priesner, 1961 and N. nevskyi Moulton, 1946, and this species is widely distributed in the southern Palearctic. In contrast, N. persica and N. sinaitica are known only from Iran and Egypt respectively, and N. gigantea from Egypt to Morocco. The following six new combinations involve species with less than three sensoria on the third antennal segment: Haplothrips biroi (Priesner, 1928), H. faurei (Zur Strassen, 1966), H. hrasvamukha (Ramakrishna, 1928), H. johni (Priesner, 1925), H. lederi (Priesner, 1924), and H. verbasci (Osborn, 1897). One new combination involves an unrelated species from India, Xylaplothrips montanus (Ananthakrishnan & Jagadish, 1970). The available biological data suggest that species of Neoheegeria are associated particularly with the flowers of Lamiaceae. K. Minaei * & P. Azemayeshfard, ����������������Plant Protection D����������epartment, F���������aculty of Horticultural Science and Plant Protection, College of Agriculture and Natural Resources, University of Tehran, Iran, [email protected] L.A. -

Pu'u Wa'awa'a Biological Assessment
PU‘U WA‘AWA‘A BIOLOGICAL ASSESSMENT PU‘U WA‘AWA‘A, NORTH KONA, HAWAII Prepared by: Jon G. Giffin Forestry & Wildlife Manager August 2003 STATE OF HAWAII DEPARTMENT OF LAND AND NATURAL RESOURCES DIVISION OF FORESTRY AND WILDLIFE TABLE OF CONTENTS TITLE PAGE ................................................................................................................................. i TABLE OF CONTENTS ............................................................................................................. ii GENERAL SETTING...................................................................................................................1 Introduction..........................................................................................................................1 Land Use Practices...............................................................................................................1 Geology..................................................................................................................................3 Lava Flows............................................................................................................................5 Lava Tubes ...........................................................................................................................5 Cinder Cones ........................................................................................................................7 Soils .......................................................................................................................................9 -

DNA Barcoding, an Approach for Molecular Identification of Huyen-Sam
LIFE SCIENCES | PHARMACOLOGY, BIOTECHNOLOGY DNA barcoding, an approach for molecular identification of Huyen-sam (Scrophularia L.) samples collected in Northern Vietnam Manh Minh Bui1, Anh Tuan Vu2, Phuong Nhung Vu1, Quang Cu Pham2, Dang Ton Nguyen1,3, Thi Thu Hue Huynh1,3* 1Institute of Genome Research, Vietnam Academy of Science and Technology 2General Department of Logistics - Techniques 3Graduate University of Science and Technology, Vietnam Academy of Science and Technology Received 4 December 2017; accepted 26 March 2018 Abstract: Introduction Huyen-sam (Vietnamese name) which belongs Scrophularia L., which is commonly called “figwort” to Scrophularia L. genus is a valuable herb. This is a plant genus belonging to family Scrophulariaceae. medicinal plant is classified as Scrophularia ningpoensis The genus comprises about 200-300 species distributed in Hemsl. Huyen-sam roots, which contain a large amount Central Asia, Europe (Mediterranean), North America and of bioactive compounds, have a similar morphology China [1-3]. Huyen-sam (Vietnamese name) or Scrophularia to its relatives. DNA barcodes promise to be a precise and reliable tool for distinguishing the processed ningpoensis Hemsl., whose root is a valuable natural herb, is Huyen-sam materials from their counterfeits. However, usually used for the treatment of inflammation, constipation studies about using DNA barcodes for classification and fever [4-6]. The main bioactive compounds present of Scrophularia L. in Vietnam are not available. in S. ningpoensis’s root are harpagoside, angroside C, Here, we conducted a taxonomic analysis of eight acteoside and cinnamic acid, which have anti-inflammatory, Scrophularia L. samples collected from the mountain antimicrobial and antioxidant effects [3, 6, 7]. -

Temporal and Spatial Differences in Gall Induction on Haloxylon by Aceria Haloxylonis (Acari: Eriophyidae) in the Gurbantünggüt Desert
Systematic & Applied Acarology 21(12): 1670–1680 (2016) ISSN 1362-1971 (print) http://doi.org/10.11158/saa.21.12.8 ISSN 2056-6069 (online) Article Temporal and spatial differences in gall induction on Haloxylon by Aceria haloxylonis (Acari: Eriophyidae) in the Gurbantünggüt Desert FEN-LIAN LI*, TING LI*, JIE SU, SHUAI YANG, PEI-LING WANG & JIAN-PING ZHANG1 Agricultural College, Shihezi University, Xinjiang, 832000, China 1Corresponding author: E–mail: [email protected] * These authors contributed equally to this work Abstract Flower-like galls have been observed on Haloxylon ammodendron and H. persicum in the Gurbantünggüt Desert in northwest China. The galls were induced by Aceria haloxylonis, a new species of Eriophyidae. The galls began as small protuberances at the base of new stems and on small branches. As they matured, the galls changed color from green to dark brown. Some galls on H. persicum became red. At maturity, the galls and the infected branches became desiccated. Adult females of A. haloxylonis overwintered in galls or in branch crevices of H. ammodendron and H. persicum. There were more galls on H. ammodendron than on H. persicum. Several ecological factors influenced gall number, including terrain, tree size, branch direction and slope aspect. H. ammodendron trees in gravel desert had more galls than trees at sand dune edges. Trees in the interdune space had the fewest galls. Large H. ammodendron trees had significantly more galls than small trees. Branches on the south side of the tree had more galls than branches on the north, east, and west sides. Terrain * tree size had significant interaction on gall number on H. -

Anti Alzheimer's Disease Related Activities of Israeli Medicinal Plants
Central JSM Alzheimer’s Disease and Related Dementia Research Article *Corresponding author Hanna Rosenmann, Department of Neurology, Hadassah University Hospital, Ein Karem, Jerusalem 91120, Anti Alzheimer’s Disease Israel, Tel: 972-2-6776954; Fax: 972-3-7256022; Email: Related Activities of Israeli Submitted: 03 December 2014 Accepted: 24 February 2015 Published: 26 February 2015 Medicinal Plants Copyright Lea Tsalkovich1, Sarah Sallon2, Helen Paavilainen2 and Hanna © 2015 Rosenmann et al. Rosenmann1* OPEN ACCESS 1Department of Neurology, Hadassah Hebrew University Medical Center, Israel 2The Louis Borick Natural Medicine Research Center, Hadassah Hebrew University Medical Center, Israel Abstract Medicinal plants may provide a potential source of therapeutic agents against Alzheimer’s-disease (AD) and other dementias. In the current study medical plants with an historical/traditional use for improving memory were evaluated for anti-AD activity. Plants were collected mainly from wild species growing in Israel and tested for their protective properties against toxicity of amyloid-beta (Aβ) and a decrease in its secretion by neuronal cells (SHSY5Y and PC12). Of 20 plants tested, 5 plants: Citrullus colocynthis, Cistanche tubulosa, Paronychia argentea, Sisymbrium irio and Origanum dayi Post, showed rescue of cells from amyloid toxicity of 205%, 177%, 42.5%, 40%, and22.7%, respectively. Citrullus colocynthis and Cistanche tubulosa also demonstrated a decreasing trend of 19% and 25%, respectively in Aβ levels secreted by cells. Two additional plants, Varthemia iphionoides and Anchusa strigosa, showed a decreasing trend in secreted amyloid levels of 32% and 34% and a decrease of 23 and 31% in γ-secretase activity respectively. These results point to a potential therapeutic use of medicinal plantssome of which has a long historical/ traditional use for improving memory and should be the basis of further investigation for neurodegenerative diseases such as AD. -

Haloxylon Ammodendron (Chenopodiaceae)
Complete Chloroplast Genome Sequence of Holoparasite Cistanche deserticola (Orobanchaceae) Reveals Gene Loss and Horizontal Gene Transfer from Its Host Haloxylon ammodendron (Chenopodiaceae) Xi Li1., Ti-Cao Zhang1., Qin Qiao1, Zhumei Ren2, Jiayuan Zhao1, Takahiro Yonezawa1, Masami Hasegawa1, M. James C Crabbe3, Jianqiang Li4*, Yang Zhong1,5* 1 Ministry of Education Key Laboratory for Biodiversity Science and Ecological Engineering, School of Life Sciences, Fudan University, Shanghai, China, 2 College of Life Science and Technology, Shanxi University, Taiyuan, China, 3 Faculty of Creative Arts, Technologies and Science, Institute of Biomedical, Environmental Science and Technology, University of Bedfordshire, Luton, United Kingdom, 4 Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China, 5 Institute of Biodiversity Science and Geobiology, Tibet University, Lhasa, China Abstract Background: The central function of chloroplasts is to carry out photosynthesis, and its gene content and structure are highly conserved across land plants. Parasitic plants, which have reduced photosynthetic ability, suffer gene losses from the chloroplast (cp) genome accompanied by the relaxation of selective constraints. Compared with the rapid rise in the number of cp genome sequences of photosynthetic organisms, there are limited data sets from parasitic plants. Principal Findings/Significance: Here we report the complete sequence of the cp genome of Cistanche deserticola,a holoparasitic desert species belonging to the family Orobanchaceae. The cp genome of C. deserticola is greatly reduced both in size (102,657 bp) and in gene content, indicating that all genes required for photosynthesis suffer from gene loss and pseudogenization, except for psbM. The striking difference from other holoparasitic plants is that it retains almost a full set of tRNA genes, and it has lower dN/dS for most genes than another close holoparasitic plant, E. -

In Wadi Allaqi, Egypt
ENVIRONMENTAL VALUATION AND MANAGEMENT OF PLANTS IN WADI ALLAQI, EGYPT FINAL REPORT IDRC OQ w W1.44 Trent University AUGUST 1998 ENVIRONMENTAL VALUATION AND-MANAGEMENT OF PLANTS IN WADI ALLAQI, EGYPT Final report Editors: Belal, A.E. , B. Leith, J. Solway and 1. Springuel Submitted To INTERNATIONAL DEVELOPMENT RESEARCH CENTRE (IDRC) CANADA File: 95-100"1/02 127-01 UNIT OF ENVIRONMENTAL STUDIES AND DEVELOPMENT, SOUTH VALLEY UNIVERSITY, ASWAN, EGYPT A-RC hf v 5 91, 5 7 By Acknowledgements The Project team of both South Valley and Trent Universities wish to thank the International Development Research Center (IDRC) Ottawa, Canada, for supporting the project with funding and for visiting the site. We also thank the staff of the IDRC Cairo Office for their assistance. This report is based upon the knowledge, hard work, and support of many people and institutions. We thank the British Council for the support they have provided in training many members of the team and UNESCO for providing support for the Allaqi project and Biosphere Reserve. We appreciate the good working relationship that we have developed with the Egyptian Environment Affairs Agency. Dr. M. Kassas of Cairo University has provided valuable intellectual direction for the project. We thank C. Fararldi who has assisted the project in numerous ways and Gordon Dickinson for writing notes on establishing the visitor center in Wadi Allaqi We wish to thank the research offices of Trent University and South Valley University. We are deeply grateful to the residents of Wadi Allaqi for their help and continued support and patience towards our project. -
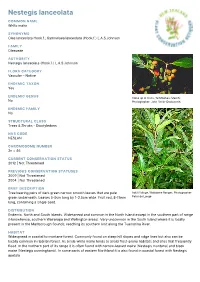
Nestegis Lanceolata
Nestegis lanceolata COMMON NAME White maire SYNONYMS Olea lanceolata Hook.f.; Gymnelaea lanceolata (Hook.f.) L.A.S.Johnson FAMILY Oleaceae AUTHORITY Nestegis lanceolata (Hook.f.) L.A.S.Johnson FLORA CATEGORY Vascular – Native ENDEMIC TAXON Yes ENDEMIC GENUS Close up of fruits, Te Moehau (March). No Photographer: John Smith-Dodsworth ENDEMIC FAMILY No STRUCTURAL CLASS Trees & Shrubs - Dicotyledons NVS CODE NESLAN CHROMOSOME NUMBER 2n = 46 CURRENT CONSERVATION STATUS 2012 | Not Threatened PREVIOUS CONSERVATION STATUSES 2009 | Not Threatened 2004 | Not Threatened BRIEF DESCRIPTION Tree bearing pairs of dark green narrow smooth leaves that are pale Adult foliage, Waitakere Ranges. Photographer: green underneath. Leaves 5-9cm long by 1-2.5cm wide. Fruit red, 8-11mm Peter de Lange long, containing a single seed. DISTRIBUTION Endemic. North and South Islands. Widespread and common in the North Island except in the southern part of range (Horowhenua, southern Wairarapa and Wellington areas). Very uncommon in the South Island where it is locally present in the Marlborough Sounds, reaching its southern limit along the Tuamarina River. HABITAT Widespread in coastal to montane forest. Commonly found on steep hill slopes and ridge lines but also can be locally common in riparian forest. As a rule white maire tends to avoid frost-prone habitats and sites that frequently flood. In the northern part of its range it is often found with narrow-leaved maire (Nestegis montana) and black maire (Nestegis cunninghamii). In some parts of eastern Northland it is also found in coastal forest with Nestegis apetala. FEATURES Stout gynodioecious spreading tree up to 20 m tall usually forming a domed canopy; trunk up to c. -

A Landscape-Based Assessment of Climate Change Vulnerability for All Native Hawaiian Plants
Technical Report HCSU-044 A LANDscape-bASED ASSESSMENT OF CLIMatE CHANGE VULNEraBILITY FOR ALL NatIVE HAWAIIAN PLANts Lucas Fortini1,2, Jonathan Price3, James Jacobi2, Adam Vorsino4, Jeff Burgett1,4, Kevin Brinck5, Fred Amidon4, Steve Miller4, Sam `Ohukani`ohi`a Gon III6, Gregory Koob7, and Eben Paxton2 1 Pacific Islands Climate Change Cooperative, Honolulu, HI 96813 2 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Hawaii National Park, HI 96718 3 Department of Geography & Environmental Studies, University of Hawai‘i at Hilo, Hilo, HI 96720 4 U.S. Fish & Wildlife Service —Ecological Services, Division of Climate Change and Strategic Habitat Management, Honolulu, HI 96850 5 Hawai‘i Cooperative Studies Unit, Pacific Island Ecosystems Research Center, Hawai‘i National Park, HI 96718 6 The Nature Conservancy, Hawai‘i Chapter, Honolulu, HI 96817 7 USDA Natural Resources Conservation Service, Hawaii/Pacific Islands Area State Office, Honolulu, HI 96850 Hawai‘i Cooperative Studies Unit University of Hawai‘i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 November 2013 This product was prepared under Cooperative Agreement CAG09AC00070 for the Pacific Island Ecosystems Research Center of the U.S. Geological Survey. Technical Report HCSU-044 A LANDSCAPE-BASED ASSESSMENT OF CLIMATE CHANGE VULNERABILITY FOR ALL NATIVE HAWAIIAN PLANTS LUCAS FORTINI1,2, JONATHAN PRICE3, JAMES JACOBI2, ADAM VORSINO4, JEFF BURGETT1,4, KEVIN BRINCK5, FRED AMIDON4, STEVE MILLER4, SAM ʽOHUKANIʽOHIʽA GON III 6, GREGORY KOOB7, AND EBEN PAXTON2 1 Pacific Islands Climate Change Cooperative, Honolulu, HI 96813 2 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Hawaiʽi National Park, HI 96718 3 Department of Geography & Environmental Studies, University of Hawaiʽi at Hilo, Hilo, HI 96720 4 U. -

A New Eriophyoid Mite Species (Acari: Eriophyidae) Infesting Haloxylon Ammodendron and H
Systematic & Applied Acarology 17(2): 202–209. ISSN 1362-1971 Article A new eriophyoid mite species (Acari: Eriophyidae) infesting Haloxylon ammodendron and H. persicum (Chenopodiaceae) in Xinjiang Uigur Autonomous Region, northwest China XIAO-FENG XUE1*, JIAN-PING ZHANG2*, FENG-LIAN LI2 & XIAO-YUE HONG1,3 1Department of Entomology, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China 2Department of Plant Protection, College of Agriculture, Shihezi University, Shihezi, Xinjiang 832003, China 3Corresponding author. E-mail: [email protected] * They contribute equally. Abstract A new mite species belonging to the Eriophyidae (Acari: Eriophyoidea) from Shihezi, Xinjiang Uigur Autonomous Region, northwestern China, is described and illustrated. The new species, Aceria haloxylonis sp. nov. causes galls on its host plants, Haloxylon ammodendron (C.A.Mey.) Bunge and Haloxylon persicum Bunge ex Boiss. et Buhse (Chenopodiaceae). Key words: Aceria haloxylonis, new species, taxonomy Introduction There are 11 species in the genus Haloxylon (Chenopodiaceae), but only H. ammodendron and H. persicum are distributed in China. Haloxylon species live mainly in deserts and can suffer drought, infertility and extreme temperature, and have salt-tolerance to some extent (Zhang 2010). In the desert ecosystem of China, they are the biggest individuals, having the highest biomass and production. The plants serve as shelter belts to impede wind erosion and stabilize sand dunes, helping counter the process of desertification. Their bark can also be a source of water. Haloxylon ammodendron is also the host of the parasitic plant Cistanche deserticola Ma (Orobanchaceae), whose fleshy stems are of medicinal importance. The genus Aceria was established by Keifer (1944) based on the type species Eriophyes tulipae Keifer, 1938. -

HAUSTORIUM 76 1 HAUSTORIUM Parasitic Plants Newsletter ISSN 1944-6969 Official Organ of the International Parasitic Plant Society (
HAUSTORIUM 76 1 HAUSTORIUM Parasitic Plants Newsletter ISSN 1944-6969 Official Organ of the International Parasitic Plant Society (http://www.parasiticplants.org/) July 2019 Number 76 CONTENTS MESSAGE FROM THE IPPS PRESIDENT (Julie Scholes)………………………………………………..………2 MEETING REPORTS 15th World Congress on Parasitic Plants, 30 June – 5 July 2019, Amsterdam, the Netherlands.………….……..2 MISTLETOE (VISCUM ALBUM) AND ITS HOSTS IN BRITAIN (Brian Spooner)……………………………10 PHELIPANCHE AEGYPTIACA IN WESTERN IRAN (Alireza Taab)……………………………………………12 NEW AND CURRENT PROJECTS Delivering high-yielding, disease-resistant finger millet to farmers…………………………………………….…..13 N2AFRICA – new Striga project – update……………………………………………………………………….…...14 Striga asiatica Madagascar fieldwork summary 2019……………………………………………………………......14 Pea (Pisum sativum) breeding for disease and pest resistance ………………………………………………….......15 REQUEST FOR SEEDS OF OROBANCHE CRENATA (Gianniantonio Domina)…………………………...…..15 PRESS REPORTS Metabolite stimulates a crop while suppressing a weed………………………………………………………….…..16 Dodder plant poses threat to trees and crops (in Kenya)………………………………………………………...….17 PhD OPPORTUNITY AT NRI (Jonne Rodenburg)…………………………………………………………………18 THESIS Sarah Huet. An overview of Phelipanche ramosa seeds: sensitivity to germination stimulants and microbiome profile. …………………………………………………………………………………………………………………..18 BOOK REVIEW Strigolactones – Biology and Applications. Ed. by Hinanit Koltai and Cristina Prandi. (Koichi Yoneyama) …………………………………………………………………………………………………………………………....19 -

Structural Diversity and Contrasted Evolution of Cytoplasmic Genomes in Flowering Plants :A Phylogenomic Approach in Oleaceae Celine Van De Paer
Structural diversity and contrasted evolution of cytoplasmic genomes in flowering plants :a phylogenomic approach in Oleaceae Celine van de Paer To cite this version: Celine van de Paer. Structural diversity and contrasted evolution of cytoplasmic genomes in flowering plants : a phylogenomic approach in Oleaceae. Vegetal Biology. Université Paul Sabatier - Toulouse III, 2017. English. NNT : 2017TOU30228. tel-02325872 HAL Id: tel-02325872 https://tel.archives-ouvertes.fr/tel-02325872 Submitted on 22 Oct 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. REMERCIEMENTS Remerciements Mes premiers remerciements s'adressent à mon directeur de thèse GUILLAUME BESNARD. Tout d'abord, merci Guillaume de m'avoir proposé ce sujet de thèse sur la famille des Oleaceae. Merci pour ton enthousiasme et ta passion pour la recherche qui m'ont véritablement portée pendant ces trois années. C'était un vrai plaisir de travailler à tes côtés. Moi qui étais focalisée sur les systèmes de reproduction chez les plantes, tu m'as ouvert à un nouveau domaine de la recherche tout aussi intéressant qui est l'évolution moléculaire (même si je suis loin de maîtriser tous les concepts...). Tu as toujours été bienveillant et à l'écoute, je t'en remercie.