Similarities Between Decapod and Insect Neuropeptidomes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Systematic and Experimental Analysis of Their Genes, Genomes, Mrnas and Proteins; and Perspective to Next Generation Sequencing
Crustaceana 92 (10) 1169-1205 CRUSTACEAN VITELLOGENIN: A SYSTEMATIC AND EXPERIMENTAL ANALYSIS OF THEIR GENES, GENOMES, MRNAS AND PROTEINS; AND PERSPECTIVE TO NEXT GENERATION SEQUENCING BY STEPHANIE JIMENEZ-GUTIERREZ1), CRISTIAN E. CADENA-CABALLERO2), CARLOS BARRIOS-HERNANDEZ3), RAUL PEREZ-GONZALEZ1), FRANCISCO MARTINEZ-PEREZ2,3) and LAURA R. JIMENEZ-GUTIERREZ1,5) 1) Sea Science Faculty, Sinaloa Autonomous University, Mazatlan, Sinaloa, 82000, Mexico 2) Coelomate Genomic Laboratory, Microbiology and Genetics Group, Industrial University of Santander, Bucaramanga, 680007, Colombia 3) Advanced Computing and a Large Scale Group, Industrial University of Santander, Bucaramanga, 680007, Colombia 4) Catedra-CONACYT, National Council for Science and Technology, CDMX, 03940, Mexico ABSTRACT Crustacean vitellogenesis is a process that involves Vitellin, produced via endoproteolysis of its precursor, which is designated as Vitellogenin (Vtg). The Vtg gene, mRNA and protein regulation involve several environmental factors and physiological processes, including gonadal maturation and moult stages, among others. Once the Vtg gene, mRNAs and protein are obtained, it is possible to establish the relationship between the elements that participate in their regulation, which could either be species-specific, or tissue-specific. This work is a systematic analysis that compares the similarities and differences of Vtg genes, mRNA and Vtg between the crustacean species reported in databases with respect to that obtained from the transcriptome of Callinectes arcuatus, C. toxotes, Penaeus stylirostris and P. vannamei obtained with MiSeq sequencing technology from Illumina. Those analyses confirm that the Vtg obtained from selected species will serve to understand the process of vitellogenesis in crustaceans that is important for fisheries and aquaculture. RESUMEN La vitelogénesis de los crustáceos es un proceso que involucra la vitelina, producida a través de la endoproteólisis de su precursor llamado Vitelogenina (Vtg). -

Redalyc.Crecimiento Del Camarón Excavador Parastacus Pugnax
Latin American Journal of Aquatic Research E-ISSN: 0718-560X [email protected] Pontificia Universidad Católica de Valparaíso Chile Ibarra, Mauricio; Arana, Patricio M. Crecimiento del camarón excavador Parastacus pugnax (Poeppig, 1835) determinado mediante técnica de marcaje Latin American Journal of Aquatic Research, vol. 39, núm. 2, julio, 2011, pp. 378-384 Pontificia Universidad Católica de Valparaíso Valparaiso, Chile Disponible en: http://www.redalyc.org/articulo.oa?id=175019398018 Cómo citar el artículo Número completo Sistema de Información Científica Más información del artículo Red de Revistas Científicas de América Latina, el Caribe, España y Portugal Página de la revista en redalyc.org Proyecto académico sin fines de lucro, desarrollado bajo la iniciativa de acceso abierto Lat. Am. J. Aquat. Res., 39(2): 378-384, 2011 Lat. Am. J. Aquat. Res. 378 DOI: 10.3856/vol39-issue2-fulltext-18 Short Communication Crecimiento del camarón excavador Parastacus pugnax (Poeppig, 1835) determinado mediante técnica de marcaje Mauricio Ibarra1 & Patricio M. Arana1 1Escuela de Ciencias del Mar, Pontificia Universidad Católica de Valparaíso P.O. Box 1020, Valparaíso, Chile RESUMEN. Para determinar el crecimiento en el camarón excavador (Parastacus pugnax) en la zona centro sur de Chile se utilizó un marbete tipo cinturón. Los parámetros longitud cefalotorácica asintótica (Lc∞) y la velocidad de incremento en longitud y peso (K), se establecieron mediante el método de Gulland & Holt (1959). El parámetro t0 se determinó mediante la ecuación inversa del modelo de von Bertalanffy, estableciéndose que las curvas de crecimiento en longitud y peso fueron definidas por los parámetros K = 0,35 -1 mm año , t0 = -0,38 años, Lc∞ = 55,9 mm y W∞ = 83,8 g, valores similares a los de Samastacus spinifrons, especie chilena de alto potencial de cultivo, y de otros parastácidos sudamericanos, tales como P. -

Full Text in Pdf Format
Vol. 25: 83–96, 2016 AQUATIC BIOLOGY Published August 24 doi: 10.3354/ab00663 Aquat Biol OPENPEN ACCESSCCESS Thermal adaptations of embryos of six terrestrial hermit crab species Katsuyuki Hamasaki1,*, Takahiro Matsuda1, Ken Takano1, Mio Sugizaki1, Yu Murakami1, Shigeki Dan2, Shuichi Kitada1 1Graduate School of Marine Science and Technology, Tokyo University of Marine Science and Technology, Minato, Konan, Tokyo 108-8477, Japan 2Research Centre for Marine Invertebrate, National Research Institute of Fisheries and Environment of Inland Sea, Fisheries Research Agency, Momoshima, Onomichi, Hiroshima 722-0061, Japan ABSTRACT: We evaluated the thermal adaptations of embryos of 6 terrestrial hermit crab species in the family Coenobitidae (genera Birgus and Coenobita): B. latro, C. brevimanus, C. cavipes, C. purpureus, C. rugosus, and C. violascens. Embryos of each species were cultured in vitro at 6 dif- ferent temperatures (18 to 34°C) in artificial seawater to avoid air desiccation; the lower threshold temperatures for embryonic development were estimated using heat summation theory equa- tions. Additionally, partial effective cumulative temperatures (> lower threshold temperature) until hatching were determined for ovigerous females of each species cultured in containers. The relationships between the embryonic growth index values (relative area of the embryonic body vs. total embryo surface) and effective cumulative temperatures were expressed using cubic equa- tions. Lower threshold temperature was estimated to be 12.7 to 14.5°C. The effective cumulative temperature and egg incubation period estimates from the appearance of the embryonic body to hatching were higher in B. latro and C. brevimanus, followed by C. rugosus, C. cavipes, and C. violascens, and lower in C. -

Nutritional Composition, Antioxidants and Antimicrobial Activities In
Research Journal of Biotechnology Vol. 15 (4) April (2020) Res. J. Biotech Nutritional Composition, Antioxidants and Antimicrobial Activities in Muscle Tissues of Mud Crab, Scylla paramamosain Wan Roslina Wan Yusof1*, Noorasmin Mokhtar Ahmad2, Mohd Alhafiizh Zailani1, Mardhiah Mohd Shahabuddin1, Ngieng Ngui Sing2 and Awang Ahmad Sallehin Awang Husaini2 1. Centre for Pre-University Studies, Universiti Malaysia Sarawak, 94300 Kota Samarahan, Sarawak, MALAYSIA 2. Faculty of Resource Science and Technology, Universiti Malaysia Sarawak, 94300 Kota Samarahan, Sarawak, MALAYSIA *[email protected] Abstract paramamosain, Scylla serrata, Scylla transquebarica and Mud crab, Scylla paramamosain known as a green mud Scylla olivacea. Interestingly, S. paramamosain also known crab, has become a popular seafood due to its meat as a green mud crab is one of the popular species and widely quality. In addition, this marine invertebrate has been distributed in mangrove area with high water salinity such as continental coast of South China Sea and Java Sea.10 found to possess peptides with different biological activities and potentials. The aim was, first, to Among the other species, S. paramamosain has triangular determine the basic nutritional content and second, to frontal lobe spines and easily identified by the dotted pattern screen for the antioxidants and antimicrobials on its propodus. From nutritional point of view, mud crabs activities in the tissue of mud crab, S. paramamosain. have high protein, minerals and polyunsaturated fatty acids Percentages of carbohydrate, protein and fat in S. contents.23 Apart from nutritional view, many studies have paramamosain were 2.32%, 12.53% and 0.23% been conducted on the biological activities in mud crabs, respectively. -

Mud Crab Susceptibility to Disease from White Spot Syndrome Virus Is Species-Dependent Somboonna Et Al
Mud crab susceptibility to disease from white spot syndrome virus is species-dependent Somboonna et al. Somboonna et al. BMC Research Notes 2010, 3:315 http://www.biomedcentral.com/1756-0500/3/315 (20 November 2010) Somboonna et al. BMC Research Notes 2010, 3:315 http://www.biomedcentral.com/1756-0500/3/315 SHORT REPORT Open Access Mud crab susceptibility to disease from white spot syndrome virus is species-dependent Naraporn Somboonna1,2,5*, Seksan Mangkalanan3, Attasit Udompetcharaporn2, Chartchai Krittanai3, Kallaya Sritunyalucksana1,2, TW Flegel1,2,4 Abstract Background: Based on a report for one species (Scylla serrata), it is widely believed that mud crabs are relatively resistant to disease caused by white spot syndrome virus (WSSV). We tested this hypothesis by determining the degree of susceptibility in two species of mud crabs, Scylla olivacea and Scylla paramamosain, both of which were identified by mitochondrial 16 S ribosomal gene analysis. We compared single-dose and serial-dose WSSV challenges on S. olivacea and S. paramamosain. Findings: In a preliminary test using S. olivacea alone, a dose of 1 × 106 WSSV copies/g gave 100% mortality within 7 days. In a subsequent test, 17 S. olivacea and 13 S. paramamosain were divided into test and control groups for challenge with WSSV at 5 incremental, biweekly doses starting from 1 × 104 and ending at 5 × 106 copies/g. For 11 S. olivacea challenged, 3 specimens died at doses between 1 × 105 and 5 × 105 copies/g and none died for 2 weeks after the subsequent dose (1 × 106 copies/g) that was lethal within 7 days in the preliminary test. -

The Effect of Different Ph and Photoperiod Regimens on The
Short communication: The effect of different pH and photoperiod regimens on the survival rate and developmental period of the larvae of Portunus pelagicus (Decapoda, Brachyura, Portunidae) Item Type article Authors Ravi, R.; Manisseri, M.K. Download date 28/09/2021 09:20:50 Link to Item http://hdl.handle.net/1834/37376 Iranian Journal of Fisheries Sciences Short Communication 12(2) 490-499 2013 __________________________________________________________________________________________ The effect of different pH and photoperiod regimens on the survival rate and developmental period of the larvae of Portunus pelagicus (Decapoda, Brachyura, Portunidae) Ravi R.*; Manisseri M. K. Received: December 2011 Accepted: August 2012 -Marine Biodiversity Division, Central Marine Fisheries Research Institute, Post Box No. 1603, Kochi-682 018, India *Corresponding author’s e mail: [email protected] Keywords: Portunus pelagicus, Photoperiod, Larval rearing, Larval development, Larval survival. Crustacean larval development occurs (Waddy and Aiken, 1991), cannibalism within a narrow range of environmental (Hecht and Pienaar, 1993) and swimming parameters (Sastry, 1983). Among the activity and hence metabolism (Gardner, abiotic characteristics that influence the 1996). developmental period and survival rate of The effects of photoperiod on the larvae, pH and photoperiod are of great larval survival have been studied in several importance. High and low levels of pH are species such as the common shrimp known to adversely affect the physiology Crangon -
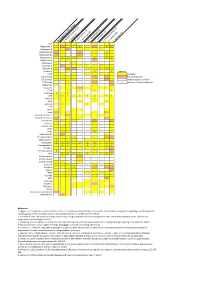
Neuropeptide Comparative Distribution.Xlsx
Allatostatin‐A 3 4 Allatostatin‐B Allatostatin‐B1 2 ACP Allatostatin‐B2 2 Allatostatin‐C 2 2 2 2 3 2 Allatostatin‐cc N. norgevicus (this study) Allatotropin C. quad ri carinatus (1) Bursicon‐A Bursicon‐B P. clark ii (2) Calcitonin 2 2 Legends CCHamide‐1H. ame Partial sequencesricanus (3) CCHamide‐2 CCAP S. verreauxi (4) CCRFamide n number of isoforms detected CNMamide M. rosenbergii (5) Corazonin CFSH‐likeCFSH 4 2 4 4 L. vann amei (6) CHH‐likeCHH 3 2 4 4 3 2 8 3 3 S. par amamosain (7) MIH‐like 2 2 Eclosion hormone 1 MIH 3 4 2 C. maenas (8) Eclosion hormone 2 E. sinensis (9) DH31 ITP DH44 C GSEFLamide C FMRFamideElevenin C C Kinin/LeucokininHIGSLYRamide 2 2 GPA2 MyosuppressinGPB5 6 2 C C Neuropeptide FNeuroparsin 2 2 3 2 3 2 3 2 3 4 2 5 3 3 Periviscerokinin 3 Orcokinin 2 Prohormone‐1 Prohormone‐3 Prohormone‐4 PDH 3 3 1 2 5 5 3 Available Undetected/not available Proctolin Pyrokinin 2 Relaxin RyamideRPCH Vasopressin‐neurophysin SIFamidesNPF 2 References Sulfakinin 1. Nguyen TV, Cummins SF, Elizur A, Ventura T. 2016. Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the Tachykinin eyestalk ganglia of the Australian crayfish, 2. Veenstra JA. 2015. The power of next‐generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. General and WXXXRamide Trissin comparative endocrinology 224: 84‐95. 3. Christie AE, Chi M, Lameyer TJ, Pascual MG, Shea DN, Stanhope ME, Schulz DJ, Dickinson PS. 2015. Neuropeptidergic signaling in the American lobster Homarus americanus 4. -

Neuropeptides in the Cerebral Ganglia of the Mud Crab, Scylla
www.nature.com/scientificreports OPEN Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic Received: 30 May 2015 Accepted: 23 October 2015 analysis and expression profiles Published: 23 November 2015 during vitellogenesis Chenchang Bao1, Yanan Yang3, Huiyang Huang1 & Haihui Ye1,2 Neuropeptides play a critical role in regulating animal reproduction. In vertebrates, GnRH, GnIH and kisspeptin are the key neuropeptide hormones of the reproductive axis, however, the reproductive axis for invertebrates is vague. Knowledge on ovarian development of the mud crab, Scylla paramamosain, is critical for aquaculture and resources management of the commercially important species. This study employed Illumina sequencing, reverse transcription-polymerase chain reaction and quantitative real-time PCR techniques to identify neuropeptides that may be involved in ovarian development of S. paramamosain. A total of 32 neuropeptide transcripts from two dozen neuropeptide families, 100 distinct mature peptides were predicted from the transcriptome data of female S. paramamosain cerebral ganglia. Among them, two families, i.e. GSEFLamide and WXXXRamide, were first identified from the cerebral ganglia of crustaceans. Of these neuropeptides, 21 transcripts of interest were selected for further confirmation and all of them were detected in the cerebral ganglia, as well as in other nervous tissues and the ovary. Most of them also had differential expression in the cerebral ganglia during various vitellogenic stages, suggesting their likely involvement in regulating vitellogenesis and ovarian maturation. Overall, these findings provide an important basis for subsequent studies on peptide function in reproduction of S. paramamosain. Neuropeptides are ubiquitous in the nervous system of the animal kingdom from cnidarians to human1. -

Reproductive Performance and Offspring Quality in Mud Crab (Scylla Paramamosain) Broodstock Fed Different Diets
Aquaculture International 11: 3—15, 2003. © 2003 Kluwer Academic Publishers. Printed in the Netherlands. Reproductive performance and offspring quality in mud crab (Scylla paramamosain) broodstock fed different diets UN S. DJUNAIDAH1*, M. WILLE2, E.K. KONTARA1 and P. SORGELOOS2 ' Center for Brackishwater Aquaculture Development, Jalan Pemandian Kartini, PO Box I, Jepara, Indonesia; 2Laboratory of Aquaculture and Artemia Reference Center, Ghent University, Rozier 44, B-9000 Ghent, Belgium Received 20 November 2001; accepted 5 October 2002 Abstract A 2-month feeding trial was conducted to evaluate the reproductive performance and offspring quality o f mud crabScylla ( paramamosain) females fed either a mixture of fresh food items (squid, shrimp, trash fish andArtemia biomass) or two experimental diets developed for penaeids. Before test initiation, mud crab females with an average individual wet weight of 200-300 g were acclimated for 2-3 days and reared together in one concrete tank of 2.0 x 0.5 x 8 m until spawning. After spawning, the spent spawners were unilaterally eyestalk ablated and randomly divided (20 animals/treatment) over three tanks of the same size and subjected to the dietary treatments. Spent spawners were used to eliminate the effect of feeding history. There were only minor differences in reproductive performance between dietary treat ments. No differences were observed in the duration of the latency period from eyestalk ablation to spawning. Fecundity was only marginally higher for the broodstock fed the control diet. Also egg quality seemed only slightly affected by the treatments. Egg hatching rates were slightly higher in crabs fed the formulated diets compared to those crabs fed the fresh diet. -

Alaska Crab Stock Enhancement and Rehabilitation / Bradley G
Workshop Proceedings March 14-16, 2006 Kodiak, Alaska Bradley G. Stevens, Editor Published by Alaska Sea Grant College Program University of Alaska Fairbanks AK-SG-06-04 Elmer E. Rasmuson Library Cataloging in Publication Data: Alaska crab stock enhancement and rehabilitation / Bradley G. Stevens, editor. – Fairbanks, Alaska : Alaska Sea Grant College Program, 2006. p. ; cm. (Alaska Sea Grant College Program, University of Alaska Fairbanks; AK-SG-06-04) Notes: “Workshop proceedings. March 14-16, 2006.” “Grant NA06OAR4170013, project A/161-01.” ISBN 1-56612-112-4 1. Crab fisheries—Congresses. I. Title. II. Stevens, Bradley Gene. III. Series: Alaska Sea Grant College Program report ; AK-SG-06-04. SH380.4.A43 2006 Credits This book is published by the Alaska Sea Grant College Program, supported by the U.S. Department of Commerce, NOAA National Sea Grant Office, grant NA06OAR4170013, project A/161-01; and by the University of Alaska Fairbanks with state funds. University of Alaska is an affirmative action/equal opportunity employer and educational institution. Sea Grant is a unique partnership with public and private sectors combining research, education, and technology transfer for public service. This national network of uni- versities meets changing environmental and economic needs of people in our coastal, ocean, and Great Lakes regions. Book design is by Jen Gunderson, cover design is by Brooks Pleninger, and copy-editing is by Sue Keller. Front cover photo by Art Sutch, adult red king crab. Back cover photos by Bradley G. Stevens. Left: Glaucothoe larva, transitional between zoea and first crab stage. Its job is to select habitat that will protect it from predation. -

Australian Entomolog
Volume 44, Part 3, 29 September 2017 THE AUSTRALIAN ENTOMOLOGIST ABN#: 15 875 103 670 The Australian Entomologist is a non-profit journal published in four parts annually by the Entomological Society of Queensland and is devoted to entomology of the Australian Region, including New Zealand, New Guinea and islands of the south-western Pacific. The journal is produced independently and subscription to the journal is not included with membership of the society. The Publications Committee Editor: Dr D.L. Hancock Assistant Editors: Dr G.B. Monteith, Dr F. Turco, Dr L. Popple, Ms S. Close. Business Manager: Dr G.B. Monteith ([email protected]) Subscriptions Subscriptions are payable in advance to the Business Manager, The Australian Entomologist, P.O. Box 537, Indooroopilly, Qld, Australia, 4068. For individuals: A$33.00 per annum in Australia. A$40.00 per annum in Asia-Pacific Region. A$45.00 per annum elsewhere. For institutions: A$37.00 per annum in Australia. A$45.00 per annum in Asia-Pacific Region. A$50.00 per annum elsewhere. Electronic Subscriptions: A$25 individuals, A$30 institutions. Please forward all overseas cheques/bank drafts in Australian currency. GST is not payable on our publication. ENTOMOLOGICAL SOCIETY OF QUEENSLAND (www.esq.org.au) Membership is open to anyone interested in Entomology. Meetings are normally held at the Ecosciences Precinct, Dutton Park, at 1.00pm on the second Tuesday of March-June and August-December each year. Meetings are announced in the Society’s News Bulletin which also contains reports of meetings, entomological notes, notices of other Society events and information on Members’ activities. -

Multifunctional Neuropeptides and Hormones in Insects and Other Invertebrates
International Journal of Molecular Sciences Review Leucokinins: Multifunctional Neuropeptides and Hormones in Insects and Other Invertebrates Dick R. Nässel 1,* and Shun-Fan Wu 2 1 Department of Zoology, Stockholm University, S-10691 Stockholm, Sweden 2 College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China; [email protected] * Correspondence: [email protected] Abstract: Leucokinins (LKs) constitute a neuropeptide family first discovered in a cockroach and later identified in numerous insects and several other invertebrates. The LK receptors are only distantly related to other known receptors. Among insects, there are many examples of species where genes encoding LKs and their receptors are absent. Furthermore, genomics has revealed that LK signaling is lacking in several of the invertebrate phyla and in vertebrates. In insects, the number and complexity of LK-expressing neurons vary, from the simple pattern in the Drosophila larva where the entire CNS has 20 neurons of 3 main types, to cockroaches with about 250 neurons of many different types. Common to all studied insects is the presence or 1–3 pairs of LK-expressing neurosecretory cells in each abdominal neuromere of the ventral nerve cord, that, at least in some insects, regulate secretion in Malpighian tubules. This review summarizes the diverse functional roles of LK signaling in insects, as well as other arthropods and mollusks. These functions include regulation of ion and water homeostasis, feeding, sleep–metabolism interactions, state-dependent memory formation, as well as modulation of gustatory sensitivity and nociception. Other functions are implied by the neuronal distribution of LK, but remain to be investigated.