Genetic and Morphological Variation in Callicarpa Japonica Thunb
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Conservation & Open Space Environment
Conservation & Open Space Environment CONSERVATION & OPEN SPACE ELEMENT INTRODUCTION Element Purpose The purpose of the Conservation & Open Space element is to promote the conservation and public use of natural resources. These planning directives should ensure that West Melbourne’s natural resources are supported and enhanced in balance with development. West Melbourne recently celebrated its 50th year as a Florida community. The character of being a small city is supported by its natural resources and open spaces. While much of the community has been developed, the undeveloped portions of the City west of Interstate 95 continue to act as a buffer between Florida’s agricultural heartland and developed Atlantic coastal communities. West Melbourne is located on the western slope of the Atlantic Coastal Ridge, very near the plateau. Elevations range from approximately 23 feet to approximately 29 feet above the National Geodetic Vertical Datum of 1929 (NGVD). The climate is subtropical, characterized by warm humid summers and dry mild winters. The average annual precipitation is 50.04 inches, mostly occurring in the rainy season from June to October. The climate has allowed tropical plant species to persist under the canopy of temperate zone trees in the hardwood areas. The City’s largest public environmental resource is Erna Nixon Park. The 52-acre park is home to a variety of native animal and vegetation. There is a large private conservation area west of I-95 which is home to wetland flora and fauna. The remainder of the City does not contain notable amounts of natural features which is due in part to the City’s agrarian roots. -

Callicarpa Dichotoma 'Duet'
U.S. National Arboretum Plant Introduction Callicarpa dichotoma 'Duet' The U.S. National Arboretum, in cooperation with Tennessee Technologi- cal University, presents 'Duet', a beautyberry with stable variegated foliage. In light shade or full sun, its lovely green and yellow leaves brighten the landscape throughout the summer season. Ease of propagation and low maintenance add to its symphony of charms. 'Duet' stars as a solo planting or as a mixed border or hedge, add- ing exciting contrast to your garden palette. Floral and Nursery Plants Research Unit U.S. Department of Agriculture, Agricultural Research Service, 3501 New York Ave., N.E., Washington, DC 20002 'Duet' Beautyberry Botanical Name: Callicarpa dichotoma 'Duet' (NA 72235; PI 651521) Family Verbenaceae Hardiness: USDA Zones 5-8 Development: A variegated sport of Callicarpa dichotoma 'Albifructus' was discov- ered at Tennessee Technological University in 2000 by Dr. Gary Bach- man and Mr. W. Edgar Davis. It was transferred to Dr. Sandra Reed for testing and evaluation in 2002. 'Duet' was released jointly by the U.S. National Arboretum and Tennessee Technological University on November 9, 2006. The cultivar name ‘Duet’ was registered in 2007 with the International Cultivar Registration Authority for unassigned genera. Significance: 'Duet' is the first stable variegated beautyberry. It was selected for its variegated foliage and tolerance to full sun. Like other Callicarpa, its disease and insect tolerant foliage make it an ideal plant for a low maintenance landscape. Description: Height and Width: 6 feet tall and 6.5 feet wide in 4 years. Habit: Deciduous, rounded shrub. Foliage: Medium green with distinct yellow margins. -

Plant Breading
SNA Research Conference Vol. 52 2007 Plant Breeding and Evaluation Tom Ranney Section Editor and Moderator Plant Breeding and Evaluation Section 326 SNA Research Conference Vol. 52 2007 New Callicarpa Species with Breeding Potential Ryan N. Contreras and John M. Ruter University of Georgia, Dept. of Horticulture, Tifton, GA 31793 [email protected] Index Words: beautyberry, species evaluation, ornamental plant breeding Significance to Industry: There is a great deal of available Callicarpa L. germplasm that has yet to be utilized by the nursery industry in the U.S. Taxa currently being evaluated are likely to have potential as breeding material or direct commercial marketability. With new breeding material and selections for introduction the number of beautyberry cultivars for use in southeastern gardens has the potential to expand greatly. Nature of Work: Callicarpa L. is a genus of ~150 species of shrubs and trees distributed throughout the world including warm-temperate and tropical America, SE Asia, Malaysia, Pacific Islands, and Australia (5) with the greatest concentration of species found in SE Asia, specifically the Philippine Islands (1). Of the New World species the highest concentration occurs in Cuba, with ~20 native species (1). There are currently four species commonly found in cultivation in the U.S.: C. americana L., C. bodinieri Lév., C. dichotoma (Lour.)K.Koch, and C. japonica Thunb. with a limited number of varieties or cultivars of each to choose from (3). Beautyberries, desired primarily for their handsome berries produced in fall, have been selected for white-fruiting varieties, finer textured varieties, increased berry production, and variegated foliage. -

Towards Resolving Lamiales Relationships
Schäferhoff et al. BMC Evolutionary Biology 2010, 10:352 http://www.biomedcentral.com/1471-2148/10/352 RESEARCH ARTICLE Open Access Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences Bastian Schäferhoff1*, Andreas Fleischmann2, Eberhard Fischer3, Dirk C Albach4, Thomas Borsch5, Günther Heubl2, Kai F Müller1 Abstract Background: In the large angiosperm order Lamiales, a diverse array of highly specialized life strategies such as carnivory, parasitism, epiphytism, and desiccation tolerance occur, and some lineages possess drastically accelerated DNA substitutional rates or miniaturized genomes. However, understanding the evolution of these phenomena in the order, and clarifying borders of and relationships among lamialean families, has been hindered by largely unresolved trees in the past. Results: Our analysis of the rapidly evolving trnK/matK, trnL-F and rps16 chloroplast regions enabled us to infer more precise phylogenetic hypotheses for the Lamiales. Relationships among the nine first-branching families in the Lamiales tree are now resolved with very strong support. Subsequent to Plocospermataceae, a clade consisting of Carlemanniaceae plus Oleaceae branches, followed by Tetrachondraceae and a newly inferred clade composed of Gesneriaceae plus Calceolariaceae, which is also supported by morphological characters. Plantaginaceae (incl. Gratioleae) and Scrophulariaceae are well separated in the backbone grade; Lamiaceae and Verbenaceae appear in distant clades, while the recently described Linderniaceae are confirmed to be monophyletic and in an isolated position. Conclusions: Confidence about deep nodes of the Lamiales tree is an important step towards understanding the evolutionary diversification of a major clade of flowering plants. The degree of resolution obtained here now provides a first opportunity to discuss the evolution of morphological and biochemical traits in Lamiales. -

AMERICAN BEAUTYBERRY (Callicarpa Americana)
ECOSYSTEM MANAGEMENT AND RESTORATION RESEARCH PROGRAM TECHNICAL REPORT EL-97-15 AMERICAN BEAUTYBERRY (Callicarpa americana) Section 7.5.8, U.S. ARMY CORPS OF ENGINEERS WILDLIFE RESOURCES MANAGEMENT MANUAL by Chester O. Martin DEPARTMENT OF THE ARMY Waterways Experiment Station, Corps of Engineers 3909 Halls Ferry Road, Vicksburg, Mississippi 39180-6199 and Sarah P. Mott Conservation Communications Vicksburg, Mississippi 39180 August 1997 Final Report Approved For Public Release; Distribution Is Unlimited EL-97-15 ^ 1997 Prepared for DEPARTMENT OF THE ARMY U.S. Army Corps of Engineers Washington, DC 20314-1000 Under EIRP Work Unit 32420 library OEC I 1 The contents of this report are not to be used for advertising, publication, or promotional purposes. Citation of trade names does not constitute an official endorsement or approval of the use of such commercial products. The findings of this report are not to be construed as an official Department of the Army position, unless so designated by other authorized documents. PRINTED ON RECYCLED PAPER ÍJ\ LIBRARY 92046874 "; ■ 11 -J - y] PREFACE This work was sponsored by the Headquarters, U.S. Army Corps of Engineers (HQUSACE), as part of the Ecosystem Management and Restoration Research Program (EMRRP), Work Unit 32420, entitled Development of U.S. Army Corps of Engineers Wildlife Resources Management Manual. Mr. Dave Mathis, CERD-C, was the EIRP Coordinator at the Directorate of Research and Development, HQUSACE. The Program Monitors for the study were Ms. Cheryl Smith, Mr. F. B. Juhle, and Mr. Forrester Einarsen, HQUSACE. The report serves as a section of the U.S. Army Corps of Engineers Wildlife Resources Management Manual. -
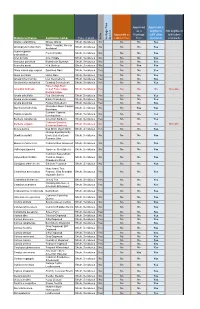
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Kadoorie Farm and Botanic Garden, 2004. Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001
Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001 Kadoorie Farm and Botanic Garden in collaboration with Zhongshan University Zhaoqing Forestry Bureau February 2004 South China Forest Biodiversity Survey Report Series: No. 37 (Online Simplified Version) Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001 Editors Bosco P.L. Chan, Ng Sai-Chit, Michael W.N. Lau and John R. Fellowes Contributors Kadoorie Farm and Botanic Garden: Michael W.N. Lau (ML) Bosco P.L. Chan (BC) John R. Fellowes (JRF) Lee Kwok Shing (LKS) Ng Sai-Chit (NSC) Roger Kendrick (RCK) Zhongshan University: Chang Hong (CH) Voluntary specialists: Graham T. Reels (GTR) Keith D.P. Wilson (KW) Background The present report details the findings of a trip to Northwest Guangdong by members of Kadoorie Farm and Botanic Garden (KFBG) in Hong Kong and their colleagues, as part of KFBG's South China Biodiversity Conservation Programme (renamed the China Programme in 2003). The overall aim of the programme is to minimise the loss of forest biodiversity in the region, and the emphasis in the first three years is on gathering up-to-date information on the distribution and status of fauna and flora. Citation Kadoorie Farm and Botanic Garden, 2004. Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001 . South China Forest Biodiversity Survey Report Series (Online Simplified Version): No. 37. KFBG, Hong Kong SAR, ii + 33 pp. Copyright Kadoorie Farm and Botanic Garden Corporation Lam Kam Road, Tai Po, N.T., Hong Kong February 2004 - i - Contents Objectives ……………………………………………………………………………………. -

Common Name Scientific Name Type Plant Family Native
Common name Scientific name Type Plant family Native region Location: Africa Rainforest Dragon Root Smilacina racemosa Herbaceous Liliaceae Oregon Native Fairy Wings Epimedium sp. Herbaceous Berberidaceae Garden Origin Golden Hakone Grass Hakonechloa macra 'Aureola' Herbaceous Poaceae Japan Heartleaf Bergenia Bergenia cordifolia Herbaceous Saxifragaceae N. Central Asia Inside Out Flower Vancouveria hexandra Herbaceous Berberidaceae Oregon Native Japanese Butterbur Petasites japonicus Herbaceous Asteraceae Japan Japanese Pachysandra Pachysandra terminalis Herbaceous Buxaceae Japan Lenten Rose Helleborus orientalis Herbaceous Ranunculaceae Greece, Asia Minor Sweet Woodruff Galium odoratum Herbaceous Rubiaceae Europe, N. Africa, W. Asia Sword Fern Polystichum munitum Herbaceous Dryopteridaceae Oregon Native David's Viburnum Viburnum davidii Shrub Caprifoliaceae Western China Evergreen Huckleberry Vaccinium ovatum Shrub Ericaceae Oregon Native Fragrant Honeysuckle Lonicera fragrantissima Shrub Caprifoliaceae Eastern China Glossy Abelia Abelia x grandiflora Shrub Caprifoliaceae Garden Origin Heavenly Bamboo Nandina domestica Shrub Berberidaceae Eastern Asia Himalayan Honeysuckle Leycesteria formosa Shrub Caprifoliaceae Himalaya, S.W. China Japanese Aralia Fatsia japonica Shrub Araliaceae Japan, Taiwan Japanese Aucuba Aucuba japonica Shrub Cornaceae Japan Kiwi Vine Actinidia chinensis Shrub Actinidiaceae China Laurustinus Viburnum tinus Shrub Caprifoliaceae Mediterranean Mexican Orange Choisya ternata Shrub Rutaceae Mexico Palmate Bamboo Sasa -

Landscape Message: Jun 12, 2020
Visit The University of Massachusetts Amherst Apply Give Search UMass.edu (/) Coronavirus (COVID-19) Resources from UMass Extension and the Center for Agriculture, Food and the Environment: ag.umass.edu/coronavirus (/coronavirus) LNUF Home (/landscape) About (/landscape/about) Newsletters & Updates (/landscape/newsletters-updates) Publications & Resources (/landscape/publications-resources) Services (/landscape/services) Education & Events (/landscape/upcoming-events) Make a Gift (https://securelb.imodules.com/s/1640/alumni/index.aspx?sid=1640&gid=2&pgid=443&cid=1121&dids=2540) Landscape Message: Jun 12, 2020 Jun 12, 2020 Issue: 9 UMass Extension's Landscape Message is an educational newsletter intended to inform and guide Massachusetts Green Industry professionals in the management of our collective landscape. Detailed (/landscape) reports from scouts and Extension specialists on growing conditions, pest activity, and cultural practices for the management of woody ornamentals, trees, and turf are regular features. The Search CAFE following issue has been updated to provide timely management information and the latest regional Search this site news and environmental data. Search UMass Extension has developed a listing of resources for commercial horticulture operations in Massachusetts to help keep you informed and aware of relevant policies and best practices for landscapers, turf managers, arborists, nurseries, garden centers, and Newsletters & greenhouse producers. Updates For our COVID-19 Information and Support for Landscapers, Nurseries, Turf Managers, Garden Centers, Arborists, and Greenhouse Operations in Massachusetts, go Landscape Message to https://ag.umass.edu/landscape/news/covid-19-information-support-for-landscapers- (/landscape/landscape-message) nurseries-turf-managers-garden-centers (/landscape/news/covid-19-information-support- Archived Landscape Messages (/landscape/landscape- for-landscapers-nurseries-turf-managers-garden-centers). -

Distribution Patterns in Malesian Callicarpa (Lamiaceae)
Gardens’ Bulletin Singapore 63(1 & 2): 287–298. 2011 287 Distribution patterns in Malesian Callicarpa (Lamiaceae) Gemma L.C. Bramley Herbarium, Library, Art and Archives, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AE, U.K. [email protected] ABSTRACT. A revision of the 55 species of Callicarpa L. (Lamiaceae) in Malesia is almost complete. There appear to be two major centres of diversity, in terms of species numbers: Borneo has 23 (44%) of the species (Bramley 2009), with 19 (83%) endemic; the Philippines has 26 (50%) of the species of which 16 (61%) are endemic (Bramley, in press, a). Callicarpa species have an extensive variation in distribution patterns; this paper focuses on the Pan- Malesian species, and the species of Borneo and the Philippines, the two islands / island groups that are the centre of Callicarpa species diversity. Fifteen of the 19 Callicarpa species endemic to Borneo belong to the ‘Geunsia’ group, an informal group used here to recognise Callicarpa pentandra and its relatives. The Geunsia group appears to be restricted to Malesia, and is only represented by C. pentandra outside of Borneo, the Philippines and Sulawesi. The 16 Callicarpa species endemic to the islands of the Philippines represent a number of different informal morphology-based groups containing species from other areas of Malesia, China or Indo-China, or, they do not appear to belong to any particular group. Keywords. Borneo, Callicarpa, centres of diversity, distribution, endemism, Malesia, Philippines Introduction A revision of Callicarpa L. (Lamiaceae) in Malesia is almost complete. It will form part of an addition and update (Bramley et al., in prep.) to Keng’s Flora Malesiana account of the Labiatae (1968). -

Lamiales – Synoptical Classification Vers
Lamiales – Synoptical classification vers. 2.6.2 (in prog.) Updated: 12 April, 2016 A Synoptical Classification of the Lamiales Version 2.6.2 (This is a working document) Compiled by Richard Olmstead With the help of: D. Albach, P. Beardsley, D. Bedigian, B. Bremer, P. Cantino, J. Chau, J. L. Clark, B. Drew, P. Garnock- Jones, S. Grose (Heydler), R. Harley, H.-D. Ihlenfeldt, B. Li, L. Lohmann, S. Mathews, L. McDade, K. Müller, E. Norman, N. O’Leary, B. Oxelman, J. Reveal, R. Scotland, J. Smith, D. Tank, E. Tripp, S. Wagstaff, E. Wallander, A. Weber, A. Wolfe, A. Wortley, N. Young, M. Zjhra, and many others [estimated 25 families, 1041 genera, and ca. 21,878 species in Lamiales] The goal of this project is to produce a working infraordinal classification of the Lamiales to genus with information on distribution and species richness. All recognized taxa will be clades; adherence to Linnaean ranks is optional. Synonymy is very incomplete (comprehensive synonymy is not a goal of the project, but could be incorporated). Although I anticipate producing a publishable version of this classification at a future date, my near- term goal is to produce a web-accessible version, which will be available to the public and which will be updated regularly through input from systematists familiar with taxa within the Lamiales. For further information on the project and to provide information for future versions, please contact R. Olmstead via email at [email protected], or by regular mail at: Department of Biology, Box 355325, University of Washington, Seattle WA 98195, USA. -

Bill Baggs Cape Florida State Park
Indian River Lagoon Preserve State Park APPROVED Unit Management Plan STATE OF FLORIDA DEPARTMENT OF ENVIRONMENTAL PROTECTION Division of Recreation and Parks April 29, 2015 TABLE OF CONTENTS INTRODUCTION ...................................................................................1 PURPOSE AND SIGNIFICANCE OF THE PARK ....................................... 1 Park Significance ................................................................................2 PURPOSE AND SCOPE OF THE PLAN..................................................... 2 MANAGEMENT PROGRAM OVERVIEW ................................................... 8 Management Authority and Responsibility .............................................. 8 Park Management Goals ...................................................................... 9 Management Coordination ................................................................... 9 Public Participation ..............................................................................9 Other Designations ........................................................................... 10 RESOURCE MANAGEMENT COMPONENT INTRODUCTION ................................................................................. 11 RESOURCE DESCRIPTION AND ASSESSMENT..................................... 12 Natural Resources ............................................................................. 12 Topography .................................................................................. 12 Geology ......................................................................................