Plant Breading
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Natural Heritage Program List of Rare Plant Species of North Carolina 2016
Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Revised February 24, 2017 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org C ur Alleghany rit Ashe Northampton Gates C uc Surry am k Stokes P d Rockingham Caswell Person Vance Warren a e P s n Hertford e qu Chowan r Granville q ot ui a Mountains Watauga Halifax m nk an Wilkes Yadkin s Mitchell Avery Forsyth Orange Guilford Franklin Bertie Alamance Durham Nash Yancey Alexander Madison Caldwell Davie Edgecombe Washington Tyrrell Iredell Martin Dare Burke Davidson Wake McDowell Randolph Chatham Wilson Buncombe Catawba Rowan Beaufort Haywood Pitt Swain Hyde Lee Lincoln Greene Rutherford Johnston Graham Henderson Jackson Cabarrus Montgomery Harnett Cleveland Wayne Polk Gaston Stanly Cherokee Macon Transylvania Lenoir Mecklenburg Moore Clay Pamlico Hoke Union d Cumberland Jones Anson on Sampson hm Duplin ic Craven Piedmont R nd tla Onslow Carteret co S Robeson Bladen Pender Sandhills Columbus New Hanover Tidewater Coastal Plain Brunswick THE COUNTIES AND PHYSIOGRAPHIC PROVINCES OF NORTH CAROLINA Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org This list is dynamic and is revised frequently as new data become available. New species are added to the list, and others are dropped from the list as appropriate. -

ANTI-MICROBIAL ACTIVITY, TOXICITY and CHEMICAL CHARACTERIZATION of EXTRACTS of Indigofera Lupatana BAKER F
ANTI-MICROBIAL ACTIVITY, TOXICITY AND CHEMICAL CHARACTERIZATION OF EXTRACTS OF Indigofera lupatana BAKER F. PLANT SOSPETER NGOCI NJERU A Thesis Submitted to the Graduate School in Partial Fulfillment for the Requirements of the Degree of Master of Science in Biochemistry of Egerton University EGERTON UNIVERSITY OCTOBER, 2010. DECLARATION AND RECOMMENDATION Declaration This thesis is my original work and has not been presented, wholly or in part, for an award of degree in any other university. Signature……………………..…………………………...Date…………………………… Sospeter Ngoci Njeru. SM14/1873/07 Recommendation We wish to confirm that this thesis was carried out under our supervision and has our approval to be presented for examination as per the Egerton University regulations. Signature………………………………………………….Date…………………………… Prof. Matasyoh J.C. Department of Chemistry Egerton University Signature……………………………………………...…Date…………………………… Dr. Mwendia C. M. Department of Biochemistry and Molecular Biology Egerton University Signature……………………………………………...…Date…………………………… Dr. Mwaniki C. G. Department of Biochemistry and Molecular Biology Egerton University ii COPYRIGHT All rights reserved. No part of this thesis may be reproduced, stored in a retrieval system or transmitted, in any form or by any means, electronic, mechanical, photocopying, or otherwise, without permission from the author. Sospeter Ngoci Njeru© 2010 iii DEDICATION This work is dedicated to my parents; Humphrey Njeru and Margaret Njeru, my wife Abishag Ngoci, my sister Flora Muthoni; Luke, Deborah and other siblings. Your constant inspiration, encouragements and support made this work successful. iv ACKNOWLEDGEMENT My sincere thanks go to Prof. Matasyoh J.C., Dr. Mwendia C. M. and Dr. Mwaniki C. G. for accepting to supervise this work and for their invaluable advice, professional insight, guidance, positive criticism, and constructive comments throughout the course of the study. -
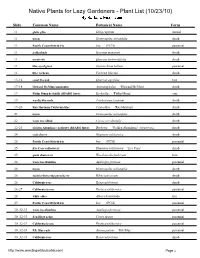
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

Nitrogen Containing Volatile Organic Compounds
DIPLOMARBEIT Titel der Diplomarbeit Nitrogen containing Volatile Organic Compounds Verfasserin Olena Bigler angestrebter akademischer Grad Magistra der Pharmazie (Mag.pharm.) Wien, 2012 Studienkennzahl lt. Studienblatt: A 996 Studienrichtung lt. Studienblatt: Pharmazie Betreuer: Univ. Prof. Mag. Dr. Gerhard Buchbauer Danksagung Vor allem lieben herzlichen Dank an meinen gütigen, optimistischen, nicht-aus-der-Ruhe-zu-bringenden Betreuer Herrn Univ. Prof. Mag. Dr. Gerhard Buchbauer ohne dessen freundlichen, fundierten Hinweisen und Ratschlägen diese Arbeit wohl niemals in der vorliegenden Form zustande gekommen wäre. Nochmals Danke, Danke, Danke. Weiteres danke ich meinen Eltern, die sich alles vom Munde abgespart haben, um mir dieses Studium der Pharmazie erst zu ermöglichen, und deren unerschütterlicher Glaube an die Fähigkeiten ihrer Tochter, mich auch dann weitermachen ließ, wenn ich mal alles hinschmeissen wollte. Auch meiner Schwester Ira gebührt Dank, auch sie war mir immer eine Stütze und Hilfe, und immer war sie da, für einen guten Rat und ein offenes Ohr. Dank auch an meinen Sohn Igor, der mit viel Verständnis akzeptierte, dass in dieser Zeit meine Prioritäten an meiner Diplomarbeit waren, und mein Zeitbudget auch für ihn eingeschränkt war. Schliesslich last, but not least - Dank auch an meinen Mann Joseph, der mich auch dann ertragen hat, wenn ich eigentlich unerträglich war. 2 Abstract This review presents a general analysis of the scienthr information about nitrogen containing volatile organic compounds (N-VOC’s) in plants. -

Bean Thrips Surveys
Blackwell Publishing AsiaMelbourne, AustraliaAENAustralian Journal of Entomology1326-6756© 2006 The Authors; Journal compilation © 2006 Australian Entomological SocietyMay 2006452122129Original ArticleSurvey for Caliothrips fasciatus in Australia M S Hoddle et al. Australian Journal of Entomology (2006) 45, 122–129 Populations of North American bean thrips, Caliothrips fasciatus (Pergande) (Thysanoptera: Thripidae: Panchaetothripinae) not detected in Australia Mark S Hoddle,1* Christina D Stosic1 and Laurence A Mound2 1Department of Entomology, University of California, Riverside, CA 92521, USA. 2Australian National Insect Collection, CSIRO Entomology, Canberra, ACT 2601, Australia. Abstract Caliothrips fasciatus is native to the USA and western Mexico and overwintering adults are regular contaminants in the ‘navel’ of navel oranges exported from California, USA to Australia, New Zealand and elsewhere. Due to the long history of regular interceptions of C. fasciatus in Australia, a survey for this thrips was undertaken around airports, seaports, public recreational parks and major agricul- tural areas in the states of Queensland, New South Wales, Victoria, South Australia and Western Australia to determine whether C. fasciatus has successfully invaded Australia. Host plants that are known to support populations of C. fasciatus, such as various annual and perennial agricultural crops, urban ornamentals and weeds along with native Australian flora, were sampled for this thrips. A total of 4675 thrips specimens encompassing at least 76 species from a minimum of 47 genera, and three families were collected from at least 159 plant species in 67 families. Caliothrips striatopterus was collected in Queensland, but the target species, C. fasciatus, was not found anywhere. An undescribed genus of Thripidae, Panchaetothripinae, was collected from ornamental Grevillea (var. -

Well-Known Plants in Each Angiosperm Order
Well-known plants in each angiosperm order This list is generally from least evolved (most ancient) to most evolved (most modern). (I’m not sure if this applies for Eudicots; I’m listing them in the same order as APG II.) The first few plants are mostly primitive pond and aquarium plants. Next is Illicium (anise tree) from Austrobaileyales, then the magnoliids (Canellales thru Piperales), then monocots (Acorales through Zingiberales), and finally eudicots (Buxales through Dipsacales). The plants before the eudicots in this list are considered basal angiosperms. This list focuses only on angiosperms and does not look at earlier plants such as mosses, ferns, and conifers. Basal angiosperms – mostly aquatic plants Unplaced in order, placed in Amborellaceae family • Amborella trichopoda – one of the most ancient flowering plants Unplaced in order, placed in Nymphaeaceae family • Water lily • Cabomba (fanwort) • Brasenia (watershield) Ceratophyllales • Hornwort Austrobaileyales • Illicium (anise tree, star anise) Basal angiosperms - magnoliids Canellales • Drimys (winter's bark) • Tasmanian pepper Laurales • Bay laurel • Cinnamon • Avocado • Sassafras • Camphor tree • Calycanthus (sweetshrub, spicebush) • Lindera (spicebush, Benjamin bush) Magnoliales • Custard-apple • Pawpaw • guanábana (soursop) • Sugar-apple or sweetsop • Cherimoya • Magnolia • Tuliptree • Michelia • Nutmeg • Clove Piperales • Black pepper • Kava • Lizard’s tail • Aristolochia (birthwort, pipevine, Dutchman's pipe) • Asarum (wild ginger) Basal angiosperms - monocots Acorales -

Vascular Flora of the Possum Walk Trail at the Infinity Science Center, Hancock County, Mississippi
The University of Southern Mississippi The Aquila Digital Community Honors Theses Honors College Spring 5-2016 Vascular Flora of the Possum Walk Trail at the Infinity Science Center, Hancock County, Mississippi Hanna M. Miller University of Southern Mississippi Follow this and additional works at: https://aquila.usm.edu/honors_theses Part of the Biodiversity Commons, and the Botany Commons Recommended Citation Miller, Hanna M., "Vascular Flora of the Possum Walk Trail at the Infinity Science Center, Hancock County, Mississippi" (2016). Honors Theses. 389. https://aquila.usm.edu/honors_theses/389 This Honors College Thesis is brought to you for free and open access by the Honors College at The Aquila Digital Community. It has been accepted for inclusion in Honors Theses by an authorized administrator of The Aquila Digital Community. For more information, please contact [email protected]. The University of Southern Mississippi Vascular Flora of the Possum Walk Trail at the Infinity Science Center, Hancock County, Mississippi by Hanna Miller A Thesis Submitted to the Honors College of The University of Southern Mississippi in Partial Fulfillment of the Requirement for the Degree of Bachelor of Science in the Department of Biological Sciences May 2016 ii Approved by _________________________________ Mac H. Alford, Ph.D., Thesis Adviser Professor of Biological Sciences _________________________________ Shiao Y. Wang, Ph.D., Chair Department of Biological Sciences _________________________________ Ellen Weinauer, Ph.D., Dean Honors College iii Abstract The North American Coastal Plain contains some of the highest plant diversity in the temperate world. However, most of the region has remained unstudied, resulting in a lack of knowledge about the unique plant communities present there. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Cytotoxicity of Aerial Parts of Indigofera Heterantha
Vol. 12(8), pp. 77-80, 30 April, 2017 DOI: 10.5897/SRE2014.5814 Article Number: 3FADC8265368 ISSN 1992-2248 Scientific Research and Essays Copyright©2017 Author(s) retain the copyright of this article http://www.academicjournals.org/SRE Full Length Research Paper Cytotoxicity of aerial parts of Indigofera heterantha Taj Ur Rahman1*, Wajiha Liaqat2, Khanzadi Fatima Khattak3, Muhammad Iqbal Choudhary4, Atif Kamil5 and Muhammad Aurang Zeb6 1Department of Chemistry, Mohi-Ud-Din Islamic University, AJ&K, Pakistan. 2Institute of Chemical Sciences, University of Peshawar, Peshawar 25120, K. P. K, Pakistan. 3Women University, Swabi, Pakistan. 4International Center for Chemical and Biological Sciences, H. E. J. Research Institute of Chemistry, University of Karachi, Karachi-75270, Pakistan. 5Department of Biotechnology Abdul Wali Khan University Mardan, Pakistan. 6Department of Biochemistry Hazara University, Mansehra, Pakistan. Received 22 January, 2014; Accepted 5 May, 2016 In the ongoing phytochemical study, an effort was made to investigate cytotoxicity of various crude fractions of aerial parts of Indigofera heterantha. The results obtained revealed that all the fractions including n-hexane, ethyl acetate, methanol and residue showed brine shrimp (Artemia salina Leach) cytotoxicity activity. The data obtained revealed the medicinal importance of the plant and will help the researchers to exploit the phytochemicals for biological activities (cytotoxicity). Key words: Indigofera heterantha, cytotoxicity, aerial parts. INTRODUCTION The genus Indigofera consists of about 700 species. All lipoxygenase inhibitory activity (Sharif et al., 2005). The species are herbs or shrubs distributed throughout the species Indigofera pulchra has shown snake-venom tropical regions of the globe. In Pakistan, this genus is neutralizing activity (Abubakar et al., 2006). -

Pages 121-166
Cost Analysis Figure 21. Estimated Unit Costs for Installation and Maintenance Procedures (2004) Costs are based on average conditions calculated from research plot applications. Costs can vary considerably depending on specific site conditions. These examples are intended for comparison purposes and should not be used as bid prices. Note: Estimated costs do not include bark mulch applied as a continuous bed. If that is the desired treatment, an additional mulch materials and application cost would apply. Estimated costs do not include plant or installation warranties. Enhancing Delaware Highways Cost Analysis 122 Figure 21. Estimated Costs for Installation and Maintenance, for comparison (2004) Drilling holes prior to planting quart containers. Note: Estimated costs do not include bark mulch applied as a continuous bed. If that is the desired treatment, an additional mulch materials and application cost would apply. Estimated costs do not include plant or installation warranties. Enhancing Delaware Highways Cost Analysis 124 Appendix A: Checklists–Inventory of Site Conditions 2. Roadway Limitations Checklist Check the roadside zone(s) included in the location to be landscaped: J Back slope or cut slope J Swale or ditch zone 1. Climate and Growth Conditions Checklist J Approach or shoulder zone J Edge or border zone J Front or fill slope Check the appropriate clear zone requirement: Check the appropriate cold hardiness zone: J Standard 30 feet J Other ( feet) J Zone 6 or J Zone 7 Presence of guard rail and/or barrier curb: Guard rail -

Towards Resolving Lamiales Relationships
Schäferhoff et al. BMC Evolutionary Biology 2010, 10:352 http://www.biomedcentral.com/1471-2148/10/352 RESEARCH ARTICLE Open Access Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences Bastian Schäferhoff1*, Andreas Fleischmann2, Eberhard Fischer3, Dirk C Albach4, Thomas Borsch5, Günther Heubl2, Kai F Müller1 Abstract Background: In the large angiosperm order Lamiales, a diverse array of highly specialized life strategies such as carnivory, parasitism, epiphytism, and desiccation tolerance occur, and some lineages possess drastically accelerated DNA substitutional rates or miniaturized genomes. However, understanding the evolution of these phenomena in the order, and clarifying borders of and relationships among lamialean families, has been hindered by largely unresolved trees in the past. Results: Our analysis of the rapidly evolving trnK/matK, trnL-F and rps16 chloroplast regions enabled us to infer more precise phylogenetic hypotheses for the Lamiales. Relationships among the nine first-branching families in the Lamiales tree are now resolved with very strong support. Subsequent to Plocospermataceae, a clade consisting of Carlemanniaceae plus Oleaceae branches, followed by Tetrachondraceae and a newly inferred clade composed of Gesneriaceae plus Calceolariaceae, which is also supported by morphological characters. Plantaginaceae (incl. Gratioleae) and Scrophulariaceae are well separated in the backbone grade; Lamiaceae and Verbenaceae appear in distant clades, while the recently described Linderniaceae are confirmed to be monophyletic and in an isolated position. Conclusions: Confidence about deep nodes of the Lamiales tree is an important step towards understanding the evolutionary diversification of a major clade of flowering plants. The degree of resolution obtained here now provides a first opportunity to discuss the evolution of morphological and biochemical traits in Lamiales. -

Indigo: Sources, Processes and Possibilities for Bioregional Blue
Indigo: Sources, processes and possibilities for bioregional blue Nicholas Wenner and Matthew Forkin November 2017 Photo by Kalie Cassel-Feiss by Kalie Photo Table of Contents Introduction . .3 Indigo . .4 The Indigo Process . 11 Conclusions . 15 Photo by Paige Green Green by Paige Photo Indigo Overview 2 Introduction his report was completed with funding generously provided by the Jena and Michael King TFoundation as part of Fibershed’s True Blue project . It is one project of many that support Fibershed’s larger mission: “Fibershed develops regional and regenerative fiber systems on behalf of independent working producers, by expanding opportunities to implement carbon farming, forming catalytic foundations to rebuild regional manufacturing, and through connecting end-users to farms and ranches through public education.” In this report we present the various sources of blue dye and of indigo, and motivate the use of plant-based indigo in particular . We also identify the limitations of natural dyes like indigo and the need for larger cultural and systemic shifts . The ideal indigo dye production system would be a closed-loop system that moves from soil to dye to textiles and back to soil . The indigo process has three basic steps: planting, harvesting, and dye extraction . In this document, we provide an overview of each, and detailed explorations are given in two separate documents that will be available through Fibershed by the end of 2017 . This report is based on a literature review of academic research, natural dye books, online content, and personal interviews . It benefited greatly from conversations with (and the generosity of) many skilled artisans and natural dyers, including Rowland Ricketts, Jane Palmer, and Kori Hargreaves .