Leader Sequence in Coupling the Stability of A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
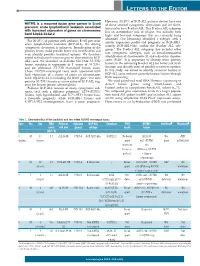
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

Transposable Elements in Human Cancers by Genome-Wide EST Alignment
Genes Genet. Syst. (2007) 82, p. 145–156 Transposable elements in human cancers by genome-wide EST alignment Dae-Soo Kim1, Jae-Won Huh2 and Heui-Soo Kim1,2* 1PBBRC, Interdisciplinary Research Program of Bioinformatics, Pusan National University, Busan 609-735, Republic of Korea 2Division of Biological Sciences, College of Natural Sciences, Pusan National University, Busan 609-735, Republic of Korea (Received 24 November 2006, accepted 23 January 2007) Transposable elements may affect coding sequences, splicing patterns, and tran- scriptional regulation of human genes. Particles of the transposable elements have been detected in several tissues and tumors. Here, we report genome-wide analysis of gene expression regulated by transposable elements in human cancers. We adopted an analysis pipeline for screening methods to detect cancer- specific expression from expressed human sequences. We developed a database (TECESdb) for understanding the mechanism of cancer development in relation to transposable elements. A total of 999 genes fused with transposable elements were found to be cancer-related in our analysis of the EST database. According to GO (Gene Ontology) analysis, the majority of the 999 cancer-specific genes have functional association with gene receptor, DNA binding, and kinase activity. Our data could contribute greatly to our understanding of human cancers in relation to transposable elements. Key words: Transposable elements, Cancer, Fusion gene, Bioinformatics, EST also appeared in open-reading frames of functional INTRODUCTION human genes (Yulug et al., 1995; Makalowski et al., 1999; The human genome is estimated to be composed of 45% Nekrutenko and Li, 2001; Huh et al., 2006). transposable elements (International Human Genome The L1 5’UTR element is known to have an antisense Sequencing Consortium 2001). -

EWSR1 Gene EWS RNA Binding Protein 1
EWSR1 gene EWS RNA binding protein 1 Normal Function The EWSR1 gene provides instructions for making the EWS protein, whose function is not completely understood. The EWS protein has two regions that contribute to its function. One region, the transcriptional activation domain, allows the EWS protein to turn on (activate) the first step in the production of proteins from genes (transcription). The other region, the RNA-binding domain, allows the EWS protein to attach (bind) to the genetic blueprint for proteins called RNA. The EWS protein may be involved in piecing together this blueprint. Some studies suggest that the RNA-binding domain is able to block (inhibit) the activity of the transcriptional activation domain, and thus regulate the function of the EWS protein. Health Conditions Related to Genetic Changes Ewing sarcoma Mutations involving the EWSR1 gene can cause a type of cancerous tumor known as Ewing sarcoma. These tumors develop in bones or soft tissues, such as nerves and cartilage. There are several types of Ewing sarcoma, including Ewing sarcoma of bone, extraosseous Ewing sarcoma, peripheral primitive neuroectodermal tumor, and Askin tumor. The mutations that cause these tumors are acquired during a person's lifetime and are present only in the tumor cells. This type of genetic change, called a somatic mutation, is not inherited. The most common mutation that causes Ewing sarcoma is a rearrangement (translocation) of genetic material between chromosome 22 and chromosome 11. This translocation, written as t(11;22), fuses part of the EWSR1 gene on chromosome 22 with part of another gene on chromosome 11 called FLI1, creating an EWSR1/FLI1 fusion gene. -

Birth of a Chimeric Primate Gene by Capture of the Transposase Gene
Birth of a chimeric primate gene by capture of the SEE COMMENTARY transposase gene from a mobile element Richard Cordaux*, Swalpa Udit†, Mark A. Batzer*, and Ce´ dric Feschotte†‡ *Department of Biological Sciences, Biological Computation and Visualization Center, Center for BioModular Multi-Scale Systems, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803; and †Department of Biology, University of Texas, Arlington, TX 76019 Edited by Susan R. Wessler, University of Georgia, Athens, GA, and approved March 27, 2006 (received for review February 10, 2006) The emergence of new genes and functions is of central impor- SETMAR transcript, which consists of these three exons, is tance to the evolution of species. The contribution of various types predicted to encode a protein of 671 amino acids and is of duplications to genetic innovation has been extensively inves- supported by 48 human cDNA clones from 18 different normal tigated. Less understood is the creation of new genes by recycling and͞or cancerous tissues (Table 1, which is published as sup- of coding material from selfish mobile genetic elements. To inves- porting information on the PNAS web site; refs. 14 and 15). tigate this process, we reconstructed the evolutionary history of These data suggest that the SETMAR protein is broadly ex- SETMAR, a new primate chimeric gene resulting from fusion of a pressed and has an important, yet unknown, function in human. SET histone methyltransferase gene to the transposase gene of a Recently, it was shown that the SET domain of the SETMAR mobile element. We show that the transposase gene was recruited protein exhibits histone methyltransferase activity (15), as do all as part of SETMAR 40–58 million years ago, after the insertion of known SET domains (16, 17). -

Selective Induction of Leukemia-Associated Fusion Genes by High-Dose Ionizing Radiation1
[CANCER RESEARCH 58. 421-425. February I. 1<W8| Selective Induction of Leukemia-associated Fusion Genes by High-Dose Ionizing Radiation1 Michael W. N. Deininger, Shikha Bose, Joanna Gora-Tybor, Xiu-Hua Yan, John M. Goldman, and Junia V. Melo2 Leukaemia Research Fumi Centre for Adult Leukaemia. Department of Haemah>li>/;\: Royal Postgraduale Medical School, Ducane Road, London W12 ONN, United Kingdom ABSTRACT event involves the acquisition of the genetic abnormality whose "success" in the production of a leukemic phenotype will depend on There is strong clinical and epidemiológica! evidence that ionizing its capacity to impart to the target cell a proliferative and/or survival radiation can cause leukemia by inducing DNA damage. This crucial advantage over its normal neighbors. In molecular terms, the gener initiation event is believed to be the result of random DNA breakage and misrepair, whereas the subsequent steps, promotion and progression, ation of a potentially successful reciprocal chromosomal translocation must rely on mechanisms of selective pressure to provide the expanding requires that: (a) at least two independent DNA DSBs occur, one in leukemic population with its proliferative/renewal advantage. To investi each chromosome partner; (b) the two breaks occur simultaneously, gate the susceptibility of human cells to external agents at the genetic i.e., within the same cell cycle, so that the two ends of one broken recombination stage of leukemogenesis, we subjected two hematopoietic chromosome are available to interact and be ligated (misrepaired) to cell lines, KG1 and III.6(1, to high doses of y-irradiation. The irradiation the respective complementary broken ends of the other chromosome; induced the formation of fusion genes characteristic of leukemia in both and (c) the recombination observes the polarity of the DNA molecule. -

DNA Transposons and the Evolution of Eukaryotic Genomes
ANRV329-GE41-15 ARI 12 October 2007 11:1 DNA Transposons and the Evolution of Eukaryotic Genomes Cedric´ Feschotte and Ellen J. Pritham Department of Biology, University of Texas, Arlington, Texas 76019; email: [email protected] Annu. Rev. Genet. 2007. 41:331–68 Key Words The Annual Review of Genetics is online at transposable elements, transposase, molecular domestication, http://genet.annualreviews.org chromosomal rearrangements This article’s doi: 10.1146/annurev.genet.40.110405.090448 Abstract Copyright c 2007 by Annual Reviews. Transposable elements are mobile genetic units that exhibit broad All rights reserved by Fordham University on 11/23/12. For personal use only. diversity in their structure and transposition mechanisms. Transpos- 0066-4197/07/1201-0331$20.00 able elements occupy a large fraction of many eukaryotic genomes and their movement and accumulation represent a major force shap- Annu. Rev. Genet. 2007.41:331-68. Downloaded from www.annualreviews.org ing the genes and genomes of almost all organisms. This review fo- cuses on DNA-mediated or class 2 transposons and emphasizes how this class of elements is distinguished from other types of mobile elements in terms of their structure, amplification dynamics, and genomic effect. We provide an up-to-date outlook on the diversity and taxonomic distribution of all major types of DNA transposons in eukaryotes, including Helitrons and Mavericks. We discuss some of the evolutionary forces that influence their maintenance and di- versification in various genomic environments. Finally, we highlight how the distinctive biological features of DNA transposons have contributed to shape genome architecture and led to the emergence of genetic innovations in different eukaryotic lineages. -

Engineering and Functional Characterization of Fusion Genes Identifies Novel Oncogenic Drivers of Cancer Hengyu Lu1, Nicole Villafane1,2, Turgut Dogruluk1, Caitlin L
Published OnlineFirst May 16, 2017; DOI: 10.1158/0008-5472.CAN-16-2745 Cancer Therapeutics, Targets, and Chemical Biology Research Engineering and Functional Characterization of Fusion Genes Identifies Novel Oncogenic Drivers of Cancer Hengyu Lu1, Nicole Villafane1,2, Turgut Dogruluk1, Caitlin L. Grzeskowiak1, Kathleen Kong1, Yiu Huen Tsang1, Oksana Zagorodna1, Angeliki Pantazi3, Lixing Yang4, Nicholas J. Neill1, Young Won Kim1, Chad J. Creighton5, Roel G. Verhaak6, Gordon B. Mills7, Peter J. Park3,4, Raju Kucherlapati3,8, and Kenneth L. Scott1,5 Abstract Oncogenic gene fusions drive many human cancers, but other reports that the transforming activity of BRAF fusions tools to more quickly unravel their functional contributions results from truncation-mediated loss of inhibitory domains are needed. Here we describe methodology permitting fusion within the N-terminus of the BRAF protein. BRAF mutations gene construction for functional evaluation. Using this strat- residing within this inhibitory region may provide a means for egy, we engineered the known fusion oncogenes, BCR-ABL1, BRAF activation in cancer, therefore we leveraged the modular EML4-ALK,andETV6-NTRK3, as well as 20 previously unchar- design of our fusion gene construction methodology to screen acterized fusion genes identifiedinTheCancerGenomeAtlas N-terminal domain mutations discovered in tumors that are datasets. In addition to confirming oncogenic activity of the wild-type at the BRAF mutation hotspot, V600. We identified known fusion oncogenes engineered by our construction strat- an oncogenic mutation, F247L, whose expression robustly egy, we validated five novel fusion genes involving MET, activated the MAPK pathway and sensitized cells to BRAF and NTRK2,andBRAF kinases that exhibited potent transforming MEK inhibitors. -

Transposon Site-Specificity and Genome Evolution
Novel insights into genome structure and evolution as a byproduct of tool generation modENCODE Symposium NHGRI Natcher Auditorium June 21, 2012 Two 20th Century surprises about the genome Transposable elements Repetitive DNA (1950) (1960) Transposons and repeats: the genomic majority Drosophila Human >30% of genome transposon-derived Full length copies Full length copies mariner 0 - 5 mariner 53,000 piggyBac 0 - 10 piggyBac 500 P element 0 - 15 P element 0* *12 Thap genes derived from P transposase The “P element”, a DNA transposon, entered genome recently (~1950), spread throughout world populations Transposons drive human evolution and cancer cell evolution But we know little about how transposons interact with the genome Hot and cold spots? Transposon-specific differences? Why do transposon-rich regions replicate late in S phase? Drosophila genome project (1991-2001: NHGRI ) and gene disruption project (2001-present: NIGMS ) PI’s: genome project- Gerry Rubin, Allan Spradling gene disruption project- Allan Spradling, Hugo Bellen, Roger Hoskins Purpose: generate insertional mutants to determine gene function of all Drosophila genes Byproduct: the best data on how transposons interact with genomes A simple experimental paradigm: Single element jumping screens: W+ W+ Advantages of this approach: Relatively unbiased Special markers to avoid silencing: yellow, rosy, Su(Var)’s Sequence flank How do you know which gene(s) are mutated? ? Association of insertion lines with genes via their insertion site requires very high quality annotation. Thank -
Consistent Rearrangement of Chromosomal Band 6P21 with Generation of Fusion Genes JAZF1/PHF1 and EPC1/PHF1 in Endometrial Stromal Sarcoma
Research Article Consistent Rearrangement of Chromosomal Band 6p21 with Generation of Fusion Genes JAZF1/PHF1 and EPC1/PHF1 in Endometrial Stromal Sarcoma Francesca Micci,1 Ioannis Panagopoulos,4 Bodil Bjerkehagen,2 and Sverre Heim1,3 Departments of 1Cancer Genetics and 2Pathology, The Norwegian Radium Hospital; 3Faculty of Medicine, University of Oslo, Oslo, Norway; and 4Department of Clinical Genetics, University Hospital, Lund, Sweden Abstract Little is known about the genetic background of ESS as only 32 Endometrial stromal sarcomas (ESS) represent <10% of all such tumors have been karyotyped and reported scientifically uterine sarcomas. Cytogenetic data on this tumor type are (6–8). The pattern of rearrangements thus detected is nevertheless limited to 32 cases, and the karyotypes are often complex, clearly nonrandom with particularly frequent involvement of but the pattern of rearrangement is nevertheless clearly chromosome arms 6p and 7p (7). Recently, a specific translocation nonrandom with particularly frequent involvement of chro- t(7;17)(p15;q21) leading to the fusion of two zinc finger genes, mosome arms 6p and 7p. Recently, a specific translocation juxtaposed with another zinc finger (JAZF1) and joined to JAZF1 t(7;17)(p15;q21) leading to the fusion of two zinc finger genes, (JJAZ1), was described in a subset of ESS (9). Both genes, the JAZF1 at 7p15 and JJAZ1 at 17q21, contain sequences encoding zinc finger juxtaposed with another zinc finger (JAZF1) and joined to JAZF1 (JJAZ1), was described in a subset of ESS. We present motifs characteristic of DNA-binding proteins. The gene fusion three ESS whose karyotypes were without the disease-specific results in expression of a tumor-specific mRNA transcript V V t(7;17) but instead showed rearrangement of chromosomal containing 5 -JAZF1 and 3 -JJAZ1 sequences but retaining the zinc band 6p21, twice as an unbalanced t(6p;7p) and once as a finger motifs from both genes. -

Comprehensive Genome and Transcriptome Analysis Reveals Genetic Basis for Gene Fusions in Cancer
bioRxiv preprint doi: https://doi.org/10.1101/148684; this version posted June 29, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Comprehensive genome and transcriptome analysis reveals genetic basis for gene fusions in cancer Nuno A. Fonseca1*, Yao He2*, Liliana Greger1, PCAWG3, Alvis Brazma1, Zemin Zhang2 1 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK; 2 Peking-Tsinghua Centre for Life Sciences, BIOPIC, and Beijing Advanced Innovation Centre for Genomics, Peking University, Beijing, 100871, China *Joint first authors Gene fusions are an important class of cancer-driving events with therapeutic and diagnostic values, yet their underlying genetic mechanisms have not been systematically characterized. Here by combining RNA and whole genome DNA sequencing data from 1188 donors across 27 cancer types we obtained a list of 3297 high-confidence tumour-specific gene fusions, 82% of which had structural variant (SV) support and 2372 of which were novel. Such a large collection of RNA and DNA alterations provides the first opportunity to systematically classify the gene fusions at a mechanistic level. While many could be explained by single SVs, numerous fusions involved series of structural rearrangements and thus are composite fusions. We discovered 75 fusions of a novel class of inter-chromosomal composite fusions, termed bridged fusions, in which a third genomic location bridged two different genes. -

Horizontal Gene Transfer in the Sponge Amphimedon Queenslandica
Horizontal gene transfer in the sponge Amphimedon queenslandica Simone Summer Higgie BEnvSc (Honours) A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2018 School of Biological Sciences Abstract Horizontal gene transfer (HGT) is the nonsexual transfer of genetic sequence across species boundaries. Historically, HGT has been assumed largely irrelevant to animal evolution, though widely recognised as an important evolutionary force in bacteria. From the recent boom in whole genome sequencing, many cases have emerged strongly supporting the occurrence of HGT in a wide range of animals. However, the extent, nature and mechanisms of HGT in animals remain poorly understood. Here, I explore these uncertainties using 576 HGTs previously reported in the genome of the demosponge Amphimedon queenslandica. The HGTs derive from bacterial, plant and fungal sources, contain a broad range of domain types, and many are differentially expressed throughout development. Some domains are highly enriched; phylogenetic analyses of the two largest groups, the Aspzincin_M35 and the PNP_UDP_1 domain groups, suggest that each results from one or few transfer events followed by post-transfer duplication. Their differential expression through development, and the conservation of domains and duplicates, together suggest that many of the HGT-derived genes are functioning in A. queenslandica. The largest group consists of aspzincins, a metallopeptidase found in bacteria and fungi, but not typically in animals. I detected aspzincins in representatives of all four of the sponge classes, suggesting that the original sponge aspzincin was transferred after sponges diverged from their last common ancestor with the Eumetazoa, but before the contemporary sponge classes emerged. -

Genes in Radiation-Induced Thyroid Carcinomas
Oncogene (1999) 18, 6330 ± 6334 ã 1999 Stockton Press All rights reserved 0950 ± 9232/99 $15.00 http://www.stockton-press.co.uk/onc Chromosomal breakpoint positions suggest a direct role for radiation in inducing illegitimate recombination between the ELE1 and RET genes in radiation-induced thyroid carcinomas YE Nikiforov*,1,2, A Koshoer1, M Nikiforova2, J Stringer3 and JA Fagin2 1Department of Pathology, University of Cincinnati College of Medicine, PO Box 670529, Cincinnati, Ohio, OH 45267-0529, USA, 2Division of Endocrinology, University of Cincinnati College of Medicine, PO Box 670547, Cincinnati, Ohio, OH 45267-0547, USA, 3Department of Molecular Genetics, University of Cincinnati College of Medicine, PO Box 670524, Cincinnati, Ohio, OH 45267-0524, USA The RET/PTC3 rearrangement is formed by fusion of Nikiforov et al., 1997), but not in tumors from the ELE1 and RET genes, and is highly prevalent in unexposed children (Bongarzone et al., 1996; Nikifor- radiation-induced post-Chernobyl papillary thyroid car- ov et al., 1997). We have also found that this type of cinomas. We characterized the breakpoints in the ELE1 RET rearrangement is associated with a speci®c and RET genes in 12 post-Chernobyl pediatric papillary histotype of radiation-induced thyroid tumors ± the carcinomas with known RET/PTC3 rearrangement. We solid growth papillary carcinoma, that is rare in found that the breakpoints within each intron were sporadic adult or pediatric populations (Nikiforov et distributed in a relatively random fashion, except for al., 1997). On the other hand, RAS and p53 mutations, clustering in the Alu regions of ELE1. None of the prevalent in sporadic thyroid cancers, are virtually breakpoints occurred at the same base or within a similar absent among post-Chernobyl tumors (Nikiforov et al., sequence.