Genome Rearrangement at Reversed-Ends Ds Element in Arabidopsis Lakshminarasimhan Krishnaswamy Iowa State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Yeast Telomeres Exert a Position Effect on Recombination Between Internal Tracts of Yeast Telomeric DNA
Downloaded from genesdev.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Yeast telomeres exert a position effect on recombination between internal tracts of yeast telomeric DNA Jeffrey B. Stavenhagen1,2 and Virginia A. Zakian Princeton University, Department of Molecular Biology, Princeton, New Jersey 08544-1014 USA; 1Biology Department, University of Dayton, Dayton, Ohio 45469-2320 USA. In Saccharomyces cerevisiae, proximity to a telomere affects both transcription and replication of adjacent DNA. In this study, we show that telomeres also impose a position effect on mitotic recombination. The rate of recombination between directly repeated tracts of telomeric C1–3A/TG1–3 DNA was reduced severely by proximity to a telomere. In contrast, recombination of two control substrates was not affected by telomere proximity. Thus, unlike position effects on transcription or replication, inhibition of recombination was sequence specific. Moreover, the repression of recombination was not under the same control as transcriptional repression (telomere position effect; TPE), as mutations in genes essential for TPE did not alleviate telomeric repression of recombination. The reduction in recombination between C1–3A/TG1–3 tracts near the telomere was caused by an absence of Rad52p-dependent events as well as a reduction in Rad1p-dependent events. The sequence-specific repression of recombination near the telomere was eliminated in cells that overexpressed the telomere-binding protein Rap1p, a condition that also increased recombination between C1–3A/TG1–3 tracts at internal positions on the chromosome. We propose that the specific inhibition between C1–3A/TG1–3 tracts near the telomere occurs through the action of a telomere-specific end-binding protein that binds to the single-strand TG1–3 tail generated during the processing of recombination intermediates. -

Genetic Effects on Microsatellite Diversity in Wild Emmer Wheat (Triticum Dicoccoides) at the Yehudiyya Microsite, Israel
Heredity (2003) 90, 150–156 & 2003 Nature Publishing Group All rights reserved 0018-067X/03 $25.00 www.nature.com/hdy Genetic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel Y-C Li1,3, T Fahima1,MSRo¨der2, VM Kirzhner1, A Beiles1, AB Korol1 and E Nevo1 1Institute of Evolution, University of Haifa, Mount Carmel, Haifa 31905, Israel; 2Institute for Plant Genetics and Crop Plant Research, Corrensstrasse 3, 06466 Gatersleben, Germany This study investigated allele size constraints and clustering, diversity. Genome B appeared to have a larger average and genetic effects on microsatellite (simple sequence repeat number (ARN), but lower variance in repeat number 2 repeat, SSR) diversity at 28 loci comprising seven types of (sARN), and smaller number of alleles per locus than genome tandem repeated dinucleotide motifs in a natural population A. SSRs with compound motifs showed larger ARN than of wild emmer wheat, Triticum dicoccoides, from a shade vs those with perfect motifs. The effects of replication slippage sun microsite in Yehudiyya, northeast of the Sea of Galilee, and recombinational effects (eg, unequal crossing over) on Israel. It was found that allele distribution at SSR loci is SSR diversity varied with SSR motifs. Ecological stresses clustered and constrained with lower or higher boundary. (sun vs shade) may affect mutational mechanisms, influen- This may imply that SSR have functional significance and cing the level of SSR diversity by both processes. natural constraints. -

Capture of a Recombination Activating Sequence from Mammalian Cells
Gene Therapy (1999) 6, 1819–1825 1999 Stockton Press All rights reserved 0969-7128/99 $15.00 http://www.stockton-press.co.uk/gt Capture of a recombination activating sequence from mammalian cells P Olson and R Dornburg The Dorrance H Hamilton Laboratories, Center for Human Virology, Division of Infectious Diseases, Jefferson Medical College, Thomas Jefferson University, Jefferson Alumni Hall, 1020 Locust Street, Rm 329, Philadelphia, PA 19107, USA We have developed a genetic trap for identifying revealed a putative recombination signal (CCCACCC). sequences that promote homologous DNA recombination. When this heptamer or an abbreviated form (CCCACC) The trap employs a retroviral vector that normally disables were reinserted into the vector, they stimulated vector itself after one round of replication. Insertion of defined repair and other DNA rearrangements. Mutant forms of DNA sequences into the vector induced the repair of a 300 these oligomers (eg CCCAACC or CCWACWS) did not. base pair deletion, which restored its ability to replicate. Our data suggest that the recombination events occurred Tests of random sequence libraries made in the vector within 48 h after transfection. Keywords: recombination; retroviral vector; vector stability; gene conversion; gene therapy Introduction scripts are still made from the intact left LTR, but reverse transcription copies the deletion to both LTRs, disabling Recognition of cis-acting DNA signals occupies a central the daughter provirus.15–17 We found that the SIN vectors role in both site-specific and general recombination path- that could escape this programmed disablement did so 1–6 ways. Signals in site-specific pathways define the by recombinationally repairing the U3-deleted LTR. -

Conversion Between 100-Million-Year-Old Duplicated Genes Contributes to Rice Subspecies Divergence
Wei et al. BMC Genomics (2021) 22:460 https://doi.org/10.1186/s12864-021-07776-y RESEARCH Open Access Conversion between 100-million-year-old duplicated genes contributes to rice subspecies divergence Chendan Wei1†, Zhenyi Wang1†, Jianyu Wang1†, Jia Teng1, Shaoqi Shen1, Qimeng Xiao1, Shoutong Bao1, Yishan Feng1, Yan Zhang1, Yuxian Li1, Sangrong Sun1, Yuanshuai Yue1, Chunyang Wu1, Yanli Wang1, Tianning Zhou1, Wenbo Xu1, Jigao Yu2,3, Li Wang1* and Jinpeng Wang1,2,3* Abstract Background: Duplicated gene pairs produced by ancient polyploidy maintain high sequence similarity over a long period of time and may result from illegitimate recombination between homeologous chromosomes. The genomes of Asian cultivated rice Oryza sativa ssp. indica (XI) and Oryza sativa ssp. japonica (GJ) have recently been updated, providing new opportunities for investigating ongoing gene conversion events and their impact on genome evolution. Results: Using comparative genomics and phylogenetic analyses, we evaluated gene conversion rates between duplicated genes produced by polyploidization 100 million years ago (mya) in GJ and XI. At least 5.19–5.77% of genes duplicated across the three rice genomes were affected by whole-gene conversion after the divergence of GJ and XI at ~ 0.4 mya, with more (7.77–9.53%) showing conversion of only portions of genes. Independently converted duplicates surviving in the genomes of different subspecies often use the same donor genes. The ongoing gene conversion frequency was higher near chromosome termini, with a single pair of homoeologous chromosomes, 11 and 12, in each rice genome being most affected. Notably, ongoing gene conversion has maintained similarity between very ancient duplicates, provided opportunities for further gene conversion, and accelerated rice divergence. -
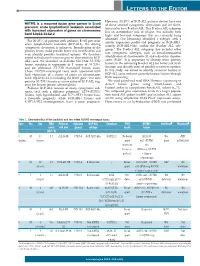
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

Genome-Wide Determination of Gene Essentiality by Transposon Insertion Sequencing in Yeast Pichia Pastoris
www.nature.com/scientificreports OPEN Genome-Wide Determination of Gene Essentiality by Transposon Insertion Sequencing in Yeast Pichia Received: 8 December 2017 Accepted: 19 June 2018 pastoris Published: xx xx xxxx Jinxiang Zhu1, Ruiqing Gong1, Qiaoyun Zhu1, Qiulin He1, Ning Xu1, Yichun Xu1, Menghao Cai1, Xiangshan Zhou1, Yuanxing Zhang1,2 & Mian Zhou1 In many prokaryotes but limited eukaryotic species, the combination of transposon mutagenesis and high-throughput sequencing has greatly accelerated the identifcation of essential genes. Here we successfully applied this technique to the methylotrophic yeast Pichia pastoris and classifed its conditionally essential/non-essential gene sets. Firstly, we showed that two DNA transposons, TcBuster and Sleeping beauty, had high transposition activities in P. pastoris. By merging their insertion libraries and performing Tn-seq, we identifed a total of 202,858 unique insertions under glucose supported growth condition. We then developed a machine learning method to classify the 5,040 annotated genes into putatively essential, putatively non-essential, ambig1 and ambig2 groups, and validated the accuracy of this classifcation model. Besides, Tn-seq was also performed under methanol supported growth condition and methanol specifc essential genes were identifed. The comparison of conditionally essential genes between glucose and methanol supported growth conditions helped to reveal potential novel targets involved in methanol metabolism and signaling. Our fndings suggest that transposon mutagenesis and Tn-seq could be applied in the methylotrophic yeast Pichia pastoris to classify conditionally essential/non-essential gene sets. Our work also shows that determining gene essentiality under diferent culture conditions could help to screen for novel functional components specifcally involved in methanol metabolism. -

A Long Polymorphic GT Microsatellite Within a Gene Promoter Mediates Non-Imprinted Allele-Specific DNA Methylation of a Cpg Island in a Goldfish Inter-Strain Hybrid
International Journal of Molecular Sciences Article A Long Polymorphic GT Microsatellite within a Gene Promoter Mediates Non-Imprinted Allele-Specific DNA Methylation of a CpG Island in a Goldfish Inter-Strain Hybrid 1,2, , 2, 2 Jianbo Zheng * y , Haomang Xu y and Huiwen Cao 1 Zhejiang Institute of Freshwater Fisheries, Huzhou 313001, China 2 College of Life Sciences, Zhejiang University, Hangzhou 310058, China * Correspondence: [email protected]; Tel.: +86-572-204-5681 These authors equally contributed to this work. y Received: 26 June 2019; Accepted: 6 August 2019; Published: 12 August 2019 Abstract: It is now widely accepted that allele-specific DNA methylation (ASM) commonly occurs at non-imprinted loci. Most of the non-imprinted ASM regions observed both within and outside of the CpG island show a strong correlation with DNA polymorphisms. However, what polymorphic cis-acting elements mediate non-imprinted ASM of the CpG island remains unclear. In this study, we investigated the impact of polymorphic GT microsatellites within the gene promoter on non-imprinted ASM of the local CpG island in goldfish. We generated various goldfish heterozygotes, in which the length of GT microsatellites or some non-repetitive sequences in the promoter of no tail alleles was different. By examining the methylation status of the downstream CpG island in these heterozygotes, we found that polymorphisms of a long GT microsatellite can lead to the ASM of the downstream CpG island during oogenesis and embryogenesis, polymorphisms of short GT microsatellites and non-repetitive sequences in the promoter exhibited no significant effect on the methylation of the CpG island. -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280 -

High-Frequency Germ Line Gene Conversion in Transgenic Mice J
MOLECULAR AND CELLULAR BIOLOGY, June 1992, p. 2545-2552 Vol. 12, No. 6 0270-7306/92/062545-08$02.00/0 Copyright ©) 1992, American Society for Microbiology High-Frequency Germ Line Gene Conversion in Transgenic Mice J. RAMANA MURTI, MICHAEL BUMBULIS, AND JOHN C. SCHIMENTI* Department of Genetics, School of Medicine, Case Western Reserve University, Cleveland, Ohio 45106 Received 8 November 1991/Accepted 28 February 1992 Gene conversion is the nonreciprocal transfer of genetic information between two related genes or DNA sequences. It can influence the evolution of gene families, having the capacity to generate both diversity and homogeneity. The potential evolutionary significance of this process is directly related to its frequency in the germ line. While measurement of meiotic inter- and intrachromosomal gene conversion frequency is routine in fungal systems, it has hitherto been impractical in mammals. We have designed a system for identifying and quantitating germ line gene conversion in mice by analyzing transgenic male gametes for a contrived recombination event. Spermatids which undergo the designed intrachromosomal gene conversion produce functional 13-galactosidase (encoded by the lacZ gene), which is visualized by histochemical staining. We observed a high incidence of lacZ-positive spermatids (-2%), which were produced by a combination of meiotic and mitotic conversion events. These results demonstrate that gene conversion in mice is an active recombinational process leading to nonparental gametic haplotypes. This high frequency of intrachromosomal gene conversion seems incompatible with the evolutionary divergence of newly duplicated genes. Hence, a process may exist to uncouple gene pairs from frequent conversion-mediated homogenization. Gene (or DNA) duplication is a major molecular mecha- aptation. -

Somatic and Germinal Recombination of a Direct Repeat in Arabidopsis
Copyright 0 1992 by the Genetics Society of America Somatic and Germinal Recombination of a Direct Repeatin Arabidopsis Farhah F. Assaad' and Ethan R. Signer Department of Biology, Massachusetts Institute of Technology, Cambridge Massachusetts 02139 Manuscript received April 10, 1992 Accepted for publication June 25, 1992 ABSTRACT Homologous recombination between a pair of directly repeated transgenes was studied in Arabi- dopsis. The test construct included two different internal, non-overlapping deletion alleles of npt (neomycin phosphotransferase) flanking an activeHPT (hygromycin phosphotransferase) gene. This construct was introduced into Arabidopsis by agrobacterium-mediated transformationwith selection for resistance to hygromycin, and two independent single-insert lines were analyzed. Selection for active NPT by resistance to kanamycin gave both fully and partly (chimeric) recombinant seedlings. Rates for one transgenic line were estimatedat <2 X 10-5 events per division for germinal and>1 0-6 events per division for somatic recombination, a much smaller difference than between meiotic and mitotic recombination in yeast. Southern analysis showed that recombinants could be formed by either crossing over or gene conversion. A surprisingly high fraction (at least 2/17) of the recombi- nants, however, appeared to result from the concerted action of two or more independent simple events. Some evolutionary implicationsare discussed. ENOMES are not static but rather in constant suggested for other organisms (HILLISet al. 1991), G dynamic flux. That is especially so in plants, can either remove a mutation from a gene copy or which lack a permanent germline and instead produce conversely allow a new mutation to spread through gametes from somatic lineages. Thus changes in the the repeat population. -

Evolutionary Dynamics of Hat DNA Transposon Families in Saccharomycetaceae
GBE Evolutionary Dynamics of hAT DNA Transposon Families in Saccharomycetaceae Ve´ronique Sarilar1,2, Claudine Bleykasten-Grosshans3,andCe´cile Neuve´glise1,2,* 1INRA, UMR 1319 Micalis, Jouy-en-Josas, France 2AgroParisTech, UMR Micalis, Jouy-en-Josas, France 3CNRS, UMR 7156, Laboratoire de Ge´ne´tique Mole´culaire, Ge´nomique et Microbiologie, Universite´ de Strasbourg, Strasbourg, France *Corresponding author: E-mail: [email protected]. Accepted: December 6, 2014 Data deposition: This project has been deposited at EMBL-ENA under the accession LM651396-LM651399. Abstract Transposable elements (TEs) are widespread in eukaryotes but uncommon in yeasts of the Saccharomycotina subphylum, in terms of both host species and genome fraction. The class II elements are especially scarce, but the hAT element Rover is a noteworthy exception that deserves further investigation. Here, we conducted a genome-wide analysis of hAT elements in 40 ascomycota. A novel family, Roamer, was found in three species, whereas Rover was detected in 15 preduplicated species from Kluyveromyces, Eremothecium,andLachancea genera, with up to 41 copies per genome. Rover acquisition seems to have occurred by horizontal transfer in a common ancestor of these genera. The detection of remote Rover copies in Naumovozyma dairenensis andinthesoleSaccharomyces cerevisiae strain AWRI1631, without synteny, suggests that two additional independent horizontal transfers took place toward these genomes. Such patchy distribution of elements prevents any anticipation of TE presence in incoming sequenced genomes, even closely related ones. The presence of both putative autonomous and defective Rover copies, as well as their diversification into five families, indicate particular dynamics of Rover elements in the Lachancea genus. -

Transposable Elements in Human Cancers by Genome-Wide EST Alignment
Genes Genet. Syst. (2007) 82, p. 145–156 Transposable elements in human cancers by genome-wide EST alignment Dae-Soo Kim1, Jae-Won Huh2 and Heui-Soo Kim1,2* 1PBBRC, Interdisciplinary Research Program of Bioinformatics, Pusan National University, Busan 609-735, Republic of Korea 2Division of Biological Sciences, College of Natural Sciences, Pusan National University, Busan 609-735, Republic of Korea (Received 24 November 2006, accepted 23 January 2007) Transposable elements may affect coding sequences, splicing patterns, and tran- scriptional regulation of human genes. Particles of the transposable elements have been detected in several tissues and tumors. Here, we report genome-wide analysis of gene expression regulated by transposable elements in human cancers. We adopted an analysis pipeline for screening methods to detect cancer- specific expression from expressed human sequences. We developed a database (TECESdb) for understanding the mechanism of cancer development in relation to transposable elements. A total of 999 genes fused with transposable elements were found to be cancer-related in our analysis of the EST database. According to GO (Gene Ontology) analysis, the majority of the 999 cancer-specific genes have functional association with gene receptor, DNA binding, and kinase activity. Our data could contribute greatly to our understanding of human cancers in relation to transposable elements. Key words: Transposable elements, Cancer, Fusion gene, Bioinformatics, EST also appeared in open-reading frames of functional INTRODUCTION human genes (Yulug et al., 1995; Makalowski et al., 1999; The human genome is estimated to be composed of 45% Nekrutenko and Li, 2001; Huh et al., 2006). transposable elements (International Human Genome The L1 5’UTR element is known to have an antisense Sequencing Consortium 2001).