Gene Transfer Into the Fungi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Morphological Re-Description of Ploeotia Pseudanisonema, and Phylogenetic Analysis of the Genus Ploeotia
A Morphological Re-Description of Ploeotia pseudanisonema, and Phylogenetic Analysis of the Genus Ploeotia by Andrew Buren Brooks (Under the direction of Mark A. Farmer) Abstract The genus Ploeotia represents a group of small, colorless, heterotrophic euglenids commonly found in shallow-water marine sediments. Species belonging to this genus have histori- cally been poorly described and studied. However, a Group I intron discovered within the small subunit ribosomal DNA gene of Ploeotia costata, makes P. costata the only euglena- zoan known to possess an actively splicing Group I intron. This intron contains conserved secondary structures that indicate monophyly with introns found in Stramenopiles and Ban- giales red algae. This project describes the internal and external morphology of a related species, Ploeotia pseudanisonema, using light and electron microscopy, and investigates the possibility of a Group I intron within P. pseudanisonema. Using recently obtained SSU rRNA sequences, we also examine the phylogeny of Ploeotia, and comment on the relationship of the genus Keelungia to Ploeotia. Index words: Ploeotia pseudanisonema, Ploeotia costata, Ploeotia vitrea, Keelungia pulex, Transmission electron microscopy, Scanning electron microscopy, Small subunit rRNA, Euglenozoan Phylogeny A Morphological Re-Description of Ploeotia pseudanisonema, and Phylogenetic Analysis of the Genus Ploeotia by Andrew Buren Brooks B.S., University of Alabama, 2009 A Thesis Submitted to the Graduate Faculty of The University of Georgia in Partial Fulfillment of the Requirements for the Degree Master of Science Athens, Georgia 2010 c 2014 Andrew Buren Brooks All Right Reserved A Morphological Re-Description of Ploeotia pseudanisonema, and Phylogenetic Analysis of the Genus Ploeotia by Andrew Buren Brooks Approved: Major Professor: Mark A. -

Medical Parasitology
MEDICAL PARASITOLOGY Anna B. Semerjyan Marina G. Susanyan Yerevan State Medical University Yerevan 2020 1 Chapter 15 Medical Parasitology. General understandings Parasitology is the study of parasites, their hosts, and the relationship between them. Medical Parasitology focuses on parasites which cause diseases in humans. Awareness and understanding about medically important parasites is necessary for proper diagnosis, prevention and treatment of parasitic diseases. The most important element in diagnosing a parasitic infection is the knowledge of the biology, or life cycle, of the parasites. Medical parasitology traditionally has included the study of three major groups of animals: 1. Parasitic protozoa (protists). 2. Parasitic worms (helminthes). 3. Arthropods that directly cause disease or act as transmitters of various pathogens. Parasitism is a form of association between organisms of different species known as symbiosis. Symbiosis means literally “living together”. Symbiosis can be between any plant, animal, or protist that is intimately associated with another organism of a different species. The most common types of symbiosis are commensalism, mutualism and parasitism. 1. Commensalism involves one-way benefit, but no harm is exerted in either direction. For example, mouth amoeba Entamoeba gingivalis, uses human for habitat (mouth cavity) and for food source without harming the host organism. 2. Mutualism is a highly interdependent association, in which both partners benefit from the relationship: two-way (mutual) benefit and no harm. Each member depends upon the other. For example, in humans’ large intestine the bacterium Escherichia coli produces the complex of vitamin B and suppresses pathogenic fungi, bacteria, while sheltering and getting nutrients in the intestine. 3. -

Mixotrophic Protists Among Marine Ciliates and Dinoflagellates: Distribution, Physiology and Ecology
FACULTY OF SCIENCE UNIVERSITY OF COPENHAGEN PhD thesis Woraporn Tarangkoon Mixotrophic Protists among Marine Ciliates and Dinoflagellates: Distribution, Physiology and Ecology Academic advisor: Associate Professor Per Juel Hansen Submitted: 29/04/10 Contents List of publications 3 Preface 4 Summary 6 Sammenfating (Danish summary) 8 สรุป (Thai summary) 10 The sections and objectives of the thesis 12 Introduction 14 1) Mixotrophy among marine planktonic protists 14 1.1) The role of light, food concentration and nutrients for 17 the growth of marine mixotrophic planktonic protists 1.2) Importance of marine mixotrophic protists in the 20 planktonic food web 2) Marine symbiont-bearing dinoflagellates 24 2.1) Occurrence of symbionts in the order Dinophysiales 24 2.2) The spatial distribution of symbiont-bearing dinoflagellates in 27 marine waters 2.3) The role of symbionts and phagotrophy in dinoflagellates with symbionts 28 3) Symbiosis and mixotrophy in the marine ciliate genus Mesodinium 30 3.1) Occurrence of symbiosis in Mesodinium spp. 30 3.2) The distribution of marine Mesodinium spp. 30 3.3) The role of symbionts and phagotrophy in marine Mesodinium rubrum 33 and Mesodinium pulex Conclusion and future perspectives 36 References 38 Paper I Paper II Paper III Appendix-Paper IV Appendix-I Lists of publications The thesis consists of the following papers, referred to in the synthesis by their roman numerals. Co-author statements are attached to the thesis (Appendix-I). Paper I Tarangkoon W, Hansen G Hansen PJ (2010) Spatial distribution of symbiont-bearing dinoflagellates in the Indian Ocean in relation to oceanographic regimes. Aquat Microb Ecol 58:197-213. -

Fungal Genomes Tell a Story of Ecological Adaptations
Folia Biologica et Oecologica 10: 9–17 (2014) Acta Universitatis Lodziensis Fungal genomes tell a story of ecological adaptations ANNA MUSZEWSKA Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawinskiego 5A, 02-106 Warsaw, Poland E-mail: [email protected] ABSTRACT One genome enables a fungus to have various lifestyles and strategies depending on environmental conditions and in the presence of specific counterparts. The nature of their interactions with other living and abiotic elements is a consequence of their osmotrophism. The ability to degrade complex compounds and especially plant biomass makes them a key component of the global carbon circulation cycle. Since the first fungal genomic sequence was published in 1996 mycology has benefited from the technolgical progress. The available data create an unprecedented opportunity to perform massive comparative studies with complex study design variants targeted at all cellular processes. KEY WORDS: fungal genomics, osmotroph, pathogenic fungi, mycorrhiza, symbiotic fungi, HGT Fungal ecology is a consequence of osmotrophy Fungi play a pivotal role both in encountered as leaf endosymbionts industry and human health (Fisher et al. (Spatafora et al. 2007). Since fungi are 2012). They are involved in biomass involved in complex relationships with degradation, plant and animal infections, other organisms, their ecological fermentation and chemical industry etc. repertoire is reflected in their genomes. They can be present in the form of The nature of their interactions with other resting spores, motile spores, amebae (in organisms and environment is defined by Cryptomycota, Blastocladiomycota, their osmotrophic lifestyle. Nutrient Chytrydiomycota), hyphae or fruiting acquisition and communication with bodies. The same fungal species symbionts and hosts are mediated by depending on environmental conditions secreted molecules. -
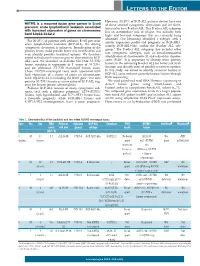
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

Transposable Elements in Human Cancers by Genome-Wide EST Alignment
Genes Genet. Syst. (2007) 82, p. 145–156 Transposable elements in human cancers by genome-wide EST alignment Dae-Soo Kim1, Jae-Won Huh2 and Heui-Soo Kim1,2* 1PBBRC, Interdisciplinary Research Program of Bioinformatics, Pusan National University, Busan 609-735, Republic of Korea 2Division of Biological Sciences, College of Natural Sciences, Pusan National University, Busan 609-735, Republic of Korea (Received 24 November 2006, accepted 23 January 2007) Transposable elements may affect coding sequences, splicing patterns, and tran- scriptional regulation of human genes. Particles of the transposable elements have been detected in several tissues and tumors. Here, we report genome-wide analysis of gene expression regulated by transposable elements in human cancers. We adopted an analysis pipeline for screening methods to detect cancer- specific expression from expressed human sequences. We developed a database (TECESdb) for understanding the mechanism of cancer development in relation to transposable elements. A total of 999 genes fused with transposable elements were found to be cancer-related in our analysis of the EST database. According to GO (Gene Ontology) analysis, the majority of the 999 cancer-specific genes have functional association with gene receptor, DNA binding, and kinase activity. Our data could contribute greatly to our understanding of human cancers in relation to transposable elements. Key words: Transposable elements, Cancer, Fusion gene, Bioinformatics, EST also appeared in open-reading frames of functional INTRODUCTION human genes (Yulug et al., 1995; Makalowski et al., 1999; The human genome is estimated to be composed of 45% Nekrutenko and Li, 2001; Huh et al., 2006). transposable elements (International Human Genome The L1 5’UTR element is known to have an antisense Sequencing Consortium 2001). -

EWSR1 Gene EWS RNA Binding Protein 1
EWSR1 gene EWS RNA binding protein 1 Normal Function The EWSR1 gene provides instructions for making the EWS protein, whose function is not completely understood. The EWS protein has two regions that contribute to its function. One region, the transcriptional activation domain, allows the EWS protein to turn on (activate) the first step in the production of proteins from genes (transcription). The other region, the RNA-binding domain, allows the EWS protein to attach (bind) to the genetic blueprint for proteins called RNA. The EWS protein may be involved in piecing together this blueprint. Some studies suggest that the RNA-binding domain is able to block (inhibit) the activity of the transcriptional activation domain, and thus regulate the function of the EWS protein. Health Conditions Related to Genetic Changes Ewing sarcoma Mutations involving the EWSR1 gene can cause a type of cancerous tumor known as Ewing sarcoma. These tumors develop in bones or soft tissues, such as nerves and cartilage. There are several types of Ewing sarcoma, including Ewing sarcoma of bone, extraosseous Ewing sarcoma, peripheral primitive neuroectodermal tumor, and Askin tumor. The mutations that cause these tumors are acquired during a person's lifetime and are present only in the tumor cells. This type of genetic change, called a somatic mutation, is not inherited. The most common mutation that causes Ewing sarcoma is a rearrangement (translocation) of genetic material between chromosome 22 and chromosome 11. This translocation, written as t(11;22), fuses part of the EWSR1 gene on chromosome 22 with part of another gene on chromosome 11 called FLI1, creating an EWSR1/FLI1 fusion gene. -

Birth of a Chimeric Primate Gene by Capture of the Transposase Gene
Birth of a chimeric primate gene by capture of the SEE COMMENTARY transposase gene from a mobile element Richard Cordaux*, Swalpa Udit†, Mark A. Batzer*, and Ce´ dric Feschotte†‡ *Department of Biological Sciences, Biological Computation and Visualization Center, Center for BioModular Multi-Scale Systems, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803; and †Department of Biology, University of Texas, Arlington, TX 76019 Edited by Susan R. Wessler, University of Georgia, Athens, GA, and approved March 27, 2006 (received for review February 10, 2006) The emergence of new genes and functions is of central impor- SETMAR transcript, which consists of these three exons, is tance to the evolution of species. The contribution of various types predicted to encode a protein of 671 amino acids and is of duplications to genetic innovation has been extensively inves- supported by 48 human cDNA clones from 18 different normal tigated. Less understood is the creation of new genes by recycling and͞or cancerous tissues (Table 1, which is published as sup- of coding material from selfish mobile genetic elements. To inves- porting information on the PNAS web site; refs. 14 and 15). tigate this process, we reconstructed the evolutionary history of These data suggest that the SETMAR protein is broadly ex- SETMAR, a new primate chimeric gene resulting from fusion of a pressed and has an important, yet unknown, function in human. SET histone methyltransferase gene to the transposase gene of a Recently, it was shown that the SET domain of the SETMAR mobile element. We show that the transposase gene was recruited protein exhibits histone methyltransferase activity (15), as do all as part of SETMAR 40–58 million years ago, after the insertion of known SET domains (16, 17). -

Selective Induction of Leukemia-Associated Fusion Genes by High-Dose Ionizing Radiation1
[CANCER RESEARCH 58. 421-425. February I. 1<W8| Selective Induction of Leukemia-associated Fusion Genes by High-Dose Ionizing Radiation1 Michael W. N. Deininger, Shikha Bose, Joanna Gora-Tybor, Xiu-Hua Yan, John M. Goldman, and Junia V. Melo2 Leukaemia Research Fumi Centre for Adult Leukaemia. Department of Haemah>li>/;\: Royal Postgraduale Medical School, Ducane Road, London W12 ONN, United Kingdom ABSTRACT event involves the acquisition of the genetic abnormality whose "success" in the production of a leukemic phenotype will depend on There is strong clinical and epidemiológica! evidence that ionizing its capacity to impart to the target cell a proliferative and/or survival radiation can cause leukemia by inducing DNA damage. This crucial advantage over its normal neighbors. In molecular terms, the gener initiation event is believed to be the result of random DNA breakage and misrepair, whereas the subsequent steps, promotion and progression, ation of a potentially successful reciprocal chromosomal translocation must rely on mechanisms of selective pressure to provide the expanding requires that: (a) at least two independent DNA DSBs occur, one in leukemic population with its proliferative/renewal advantage. To investi each chromosome partner; (b) the two breaks occur simultaneously, gate the susceptibility of human cells to external agents at the genetic i.e., within the same cell cycle, so that the two ends of one broken recombination stage of leukemogenesis, we subjected two hematopoietic chromosome are available to interact and be ligated (misrepaired) to cell lines, KG1 and III.6(1, to high doses of y-irradiation. The irradiation the respective complementary broken ends of the other chromosome; induced the formation of fusion genes characteristic of leukemia in both and (c) the recombination observes the polarity of the DNA molecule. -

DNA Transposons and the Evolution of Eukaryotic Genomes
ANRV329-GE41-15 ARI 12 October 2007 11:1 DNA Transposons and the Evolution of Eukaryotic Genomes Cedric´ Feschotte and Ellen J. Pritham Department of Biology, University of Texas, Arlington, Texas 76019; email: [email protected] Annu. Rev. Genet. 2007. 41:331–68 Key Words The Annual Review of Genetics is online at transposable elements, transposase, molecular domestication, http://genet.annualreviews.org chromosomal rearrangements This article’s doi: 10.1146/annurev.genet.40.110405.090448 Abstract Copyright c 2007 by Annual Reviews. Transposable elements are mobile genetic units that exhibit broad All rights reserved by Fordham University on 11/23/12. For personal use only. diversity in their structure and transposition mechanisms. Transpos- 0066-4197/07/1201-0331$20.00 able elements occupy a large fraction of many eukaryotic genomes and their movement and accumulation represent a major force shap- Annu. Rev. Genet. 2007.41:331-68. Downloaded from www.annualreviews.org ing the genes and genomes of almost all organisms. This review fo- cuses on DNA-mediated or class 2 transposons and emphasizes how this class of elements is distinguished from other types of mobile elements in terms of their structure, amplification dynamics, and genomic effect. We provide an up-to-date outlook on the diversity and taxonomic distribution of all major types of DNA transposons in eukaryotes, including Helitrons and Mavericks. We discuss some of the evolutionary forces that influence their maintenance and di- versification in various genomic environments. Finally, we highlight how the distinctive biological features of DNA transposons have contributed to shape genome architecture and led to the emergence of genetic innovations in different eukaryotic lineages. -

Horizontal Gene Transfer in Osmotrophs: Playing with Public Goods
ORE Open Research Exeter TITLE Horizontal gene transfer in osmotrophs: playing with public goods. AUTHORS Richards, Thomas A; Talbot, Nicholas J. JOURNAL Nat Rev Microbiol DEPOSITED IN ORE 19 November 2014 This version available at http://hdl.handle.net/10871/15898 COPYRIGHT AND REUSE Open Research Exeter makes this work available in accordance with publisher policies. A NOTE ON VERSIONS The version presented here may differ from the published version. If citing, you are advised to consult the published version for pagination, volume/issue and date of publication PERSPECTIVES however, not unique to fungi. Many bacteria, OPINION for instance, feed in an analogous man ner, and other eukaryotic groups, such as Horizontal gene transfer in hyphochytriomycetes (FIG. 1c) and oomycetes (FIG. 1d) (sometimes collectively termed the pseudofungi26), also feed osmotrophically osmotrophs: playing with public and adopt filamentous growth habits, allow ing invasive growth in heterogeneous sub goods strates. Importantly, these eukaryotes also lost the ability to carry out phagotrophy and Thomas A. Richards and Nicholas J. Talbot became obligately osmotrophic26,27. Osmotrophy has a number of distinct Abstract | Osmotrophic microorganisms, such as fungi and oomycetes, feed advantages as a feeding strategy. External by secreting depolymerizing enzymes to process complex food sources in the digestion of large and complex polymers extracellular environment, and taking up the resulting simple sugars, micronutrients allows greater control over substances that and amino acids. As a consequence of this lifestyle, osmotrophs engage in the are allowed to enter a cell (FIG. 2a), thus acquisition and protection of public goods. In this Opinion article, we propose that minimizing potential routes of infection and intake of harmful substances. -

Engineering and Functional Characterization of Fusion Genes Identifies Novel Oncogenic Drivers of Cancer Hengyu Lu1, Nicole Villafane1,2, Turgut Dogruluk1, Caitlin L
Published OnlineFirst May 16, 2017; DOI: 10.1158/0008-5472.CAN-16-2745 Cancer Therapeutics, Targets, and Chemical Biology Research Engineering and Functional Characterization of Fusion Genes Identifies Novel Oncogenic Drivers of Cancer Hengyu Lu1, Nicole Villafane1,2, Turgut Dogruluk1, Caitlin L. Grzeskowiak1, Kathleen Kong1, Yiu Huen Tsang1, Oksana Zagorodna1, Angeliki Pantazi3, Lixing Yang4, Nicholas J. Neill1, Young Won Kim1, Chad J. Creighton5, Roel G. Verhaak6, Gordon B. Mills7, Peter J. Park3,4, Raju Kucherlapati3,8, and Kenneth L. Scott1,5 Abstract Oncogenic gene fusions drive many human cancers, but other reports that the transforming activity of BRAF fusions tools to more quickly unravel their functional contributions results from truncation-mediated loss of inhibitory domains are needed. Here we describe methodology permitting fusion within the N-terminus of the BRAF protein. BRAF mutations gene construction for functional evaluation. Using this strat- residing within this inhibitory region may provide a means for egy, we engineered the known fusion oncogenes, BCR-ABL1, BRAF activation in cancer, therefore we leveraged the modular EML4-ALK,andETV6-NTRK3, as well as 20 previously unchar- design of our fusion gene construction methodology to screen acterized fusion genes identifiedinTheCancerGenomeAtlas N-terminal domain mutations discovered in tumors that are datasets. In addition to confirming oncogenic activity of the wild-type at the BRAF mutation hotspot, V600. We identified known fusion oncogenes engineered by our construction strat- an oncogenic mutation, F247L, whose expression robustly egy, we validated five novel fusion genes involving MET, activated the MAPK pathway and sensitized cells to BRAF and NTRK2,andBRAF kinases that exhibited potent transforming MEK inhibitors.