NSCLC Mutated Isoforms of CCDC6 Affect the Intracellular Distribution Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Influencers on Thyroid Cancer Onset: Molecular Genetic Basis
G C A T T A C G G C A T genes Review Influencers on Thyroid Cancer Onset: Molecular Genetic Basis Berta Luzón-Toro 1,2, Raquel María Fernández 1,2, Leticia Villalba-Benito 1,2, Ana Torroglosa 1,2, Guillermo Antiñolo 1,2 and Salud Borrego 1,2,* 1 Department of Maternofetal Medicine, Genetics and Reproduction, Institute of Biomedicine of Seville (IBIS), University Hospital Virgen del Rocío/CSIC/University of Seville, 41013 Seville, Spain; [email protected] (B.L.-T.); [email protected] (R.M.F.); [email protected] (L.V.-B.); [email protected] (A.T.); [email protected] (G.A.) 2 Centre for Biomedical Network Research on Rare Diseases (CIBERER), 41013 Seville, Spain * Correspondence: [email protected]; Tel.: +34-955-012641 Received: 3 September 2019; Accepted: 6 November 2019; Published: 8 November 2019 Abstract: Thyroid cancer, a cancerous tumor or growth located within the thyroid gland, is the most common endocrine cancer. It is one of the few cancers whereby incidence rates have increased in recent years. It occurs in all age groups, from children through to seniors. Most studies are focused on dissecting its genetic basis, since our current knowledge of the genetic background of the different forms of thyroid cancer is far from complete, which poses a challenge for diagnosis and prognosis of the disease. In this review, we describe prevailing advances and update our understanding of the molecular genetics of thyroid cancer, focusing on the main genes related with the pathology, including the different noncoding RNAs associated with the disease. -

Genome-Wide Analysis of Host-Chromosome Binding Sites For
Lu et al. Virology Journal 2010, 7:262 http://www.virologyj.com/content/7/1/262 RESEARCH Open Access Genome-wide analysis of host-chromosome binding sites for Epstein-Barr Virus Nuclear Antigen 1 (EBNA1) Fang Lu1, Priyankara Wikramasinghe1, Julie Norseen1,2, Kevin Tsai1, Pu Wang1, Louise Showe1, Ramana V Davuluri1, Paul M Lieberman1* Abstract The Epstein-Barr Virus (EBV) Nuclear Antigen 1 (EBNA1) protein is required for the establishment of EBV latent infection in proliferating B-lymphocytes. EBNA1 is a multifunctional DNA-binding protein that stimulates DNA replication at the viral origin of plasmid replication (OriP), regulates transcription of viral and cellular genes, and tethers the viral episome to the cellular chromosome. EBNA1 also provides a survival function to B-lymphocytes, potentially through its ability to alter cellular gene expression. To better understand these various functions of EBNA1, we performed a genome-wide analysis of the viral and cellular DNA sites associated with EBNA1 protein in a latently infected Burkitt lymphoma B-cell line. Chromatin-immunoprecipitation (ChIP) combined with massively parallel deep-sequencing (ChIP-Seq) was used to identify cellular sites bound by EBNA1. Sites identified by ChIP- Seq were validated by conventional real-time PCR, and ChIP-Seq provided quantitative, high-resolution detection of the known EBNA1 binding sites on the EBV genome at OriP and Qp. We identified at least one cluster of unusually high-affinity EBNA1 binding sites on chromosome 11, between the divergent FAM55 D and FAM55B genes. A con- sensus for all cellular EBNA1 binding sites is distinct from those derived from the known viral binding sites, sug- gesting that some of these sites are indirectly bound by EBNA1. -

RET/PTC Activation in Papillary Thyroid Carcinoma
European Journal of Endocrinology (2006) 155 645–653 ISSN 0804-4643 INVITED REVIEW RET/PTC activation in papillary thyroid carcinoma: European Journal of Endocrinology Prize Lecture Massimo Santoro1, Rosa Marina Melillo1 and Alfredo Fusco1,2 1Istituto di Endocrinologia ed Oncologia Sperimentale del CNR ‘G. Salvatore’, c/o Dipartimento di Biologia e Patologia Cellulare e Molecolare, University ‘Federico II’, Via S. Pansini, 5, 80131 Naples, Italy and 2NOGEC (Naples Oncogenomic Center)–CEINGE, Biotecnologie Avanzate & SEMM, European School of Molecular Medicine, Naples, Italy (Correspondence should be addressed to M Santoro; Email: [email protected]) Abstract Papillary thyroid carcinoma (PTC) is frequently associated with RET gene rearrangements that generate the so-called RET/PTC oncogenes. In this review, we examine the data about the mechanisms of thyroid cell transformation, activation of downstream signal transduction pathways and modulation of gene expression induced by RET/PTC. These findings have advanced our understanding of the processes underlying PTC formation and provide the basis for novel therapeutic approaches to this disease. European Journal of Endocrinology 155 645–653 RET/PTC rearrangements in papillary growth factor, have been described in a fraction of PTC thyroid carcinoma patients (7). As illustrated in figure 1, many different genes have been found to be rearranged with RET in The rearranged during tansfection (RET) proto-onco- individual PTC patients. RET/PTC1 and 3 account for gene, located on chromosome 10q11.2, was isolated in more than 90% of all rearrangements and are hence, by 1985 and shown to be activated by a DNA rearrange- far, the most frequent variants (8–11). They result from ment (rearranged during transfection) (1).As the fusion of RET to the coiled-coil domain containing illustrated in Fig. -

RET Gene Fusions in Malignancies of the Thyroid and Other Tissues
G C A T T A C G G C A T genes Review RET Gene Fusions in Malignancies of the Thyroid and Other Tissues Massimo Santoro 1,*, Marialuisa Moccia 1, Giorgia Federico 1 and Francesca Carlomagno 1,2 1 Department of Molecular Medicine and Medical Biotechnology, University of Naples “Federico II”, 80131 Naples, Italy; [email protected] (M.M.); [email protected] (G.F.); [email protected] (F.C.) 2 Institute of Endocrinology and Experimental Oncology of the CNR, 80131 Naples, Italy * Correspondence: [email protected] Received: 10 March 2020; Accepted: 12 April 2020; Published: 15 April 2020 Abstract: Following the identification of the BCR-ABL1 (Breakpoint Cluster Region-ABelson murine Leukemia) fusion in chronic myelogenous leukemia, gene fusions generating chimeric oncoproteins have been recognized as common genomic structural variations in human malignancies. This is, in particular, a frequent mechanism in the oncogenic conversion of protein kinases. Gene fusion was the first mechanism identified for the oncogenic activation of the receptor tyrosine kinase RET (REarranged during Transfection), initially discovered in papillary thyroid carcinoma (PTC). More recently, the advent of highly sensitive massive parallel (next generation sequencing, NGS) sequencing of tumor DNA or cell-free (cfDNA) circulating tumor DNA, allowed for the detection of RET fusions in many other solid and hematopoietic malignancies. This review summarizes the role of RET fusions in the pathogenesis of human cancer. Keywords: kinase; tyrosine kinase inhibitor; targeted therapy; thyroid cancer 1. The RET Receptor RET (REarranged during Transfection) was initially isolated as a rearranged oncoprotein upon the transfection of a human lymphoma DNA [1]. -

RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4,871 Patients Shumei Kato1, Vivek Subbiah2, Erica Marchlik3, Sheryl K
Published OnlineFirst September 28, 2016; DOI: 10.1158/1078-0432.CCR-16-1679 Personalized Medicine and Imaging Clinical Cancer Research RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4,871 Patients Shumei Kato1, Vivek Subbiah2, Erica Marchlik3, Sheryl K. Elkin3, Jennifer L. Carter3, and Razelle Kurzrock1 Abstract Purpose: Aberrations in genetic sequences encoding the tyrosine (52/88)], cell cycle–associated genes [39.8% (35/88)], the PI3K kinase receptor RET lead to oncogenic signaling that is targetable signaling pathway [30.7% (27/88)], MAPK effectors [22.7% with anti-RET multikinase inhibitors. Understanding the compre- (20/88)], or other tyrosine kinase families [21.6% (19/88)]. hensive genomic landscape of RET aberrations across multiple RET fusions were mutually exclusive with MAPK signaling cancers may facilitate clinical trial development targeting RET. pathway alterations. All 72 patients harboring coaberrations Experimental Design: We interrogated the molecular portfolio had distinct genomic portfolios, and most [98.6% (71/72)] of 4,871 patients with diverse malignancies for the presence of had potentially targetable coaberrations with either an FDA- RET aberrations using Clinical Laboratory Improvement Amend- approved or an investigational agent. Two cases with lung ments–certified targeted next-generation sequencing of 182 or (KIF5B-RET) and medullary thyroid carcinoma (RET M918T) 236 gene panels. thatrespondedtoavandetanib(multikinase RET inhibitor)- Results: Among diverse cancers, RET aberrations were iden- containing regimen are shown. tified in 88 cases [1.8% (88/4, 871)], with mutations being Conclusions: RET aberrations were seen in 1.8% of diverse the most common alteration [38.6% (34/88)], followed cancers, with most cases harboring actionable, albeit dis- by fusions [30.7% (27/88), including a novel SQSTM1-RET] tinct, coexisting alterations. -
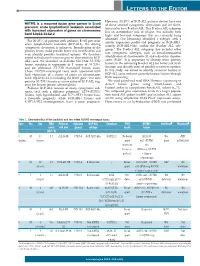
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

Transposable Elements in Human Cancers by Genome-Wide EST Alignment
Genes Genet. Syst. (2007) 82, p. 145–156 Transposable elements in human cancers by genome-wide EST alignment Dae-Soo Kim1, Jae-Won Huh2 and Heui-Soo Kim1,2* 1PBBRC, Interdisciplinary Research Program of Bioinformatics, Pusan National University, Busan 609-735, Republic of Korea 2Division of Biological Sciences, College of Natural Sciences, Pusan National University, Busan 609-735, Republic of Korea (Received 24 November 2006, accepted 23 January 2007) Transposable elements may affect coding sequences, splicing patterns, and tran- scriptional regulation of human genes. Particles of the transposable elements have been detected in several tissues and tumors. Here, we report genome-wide analysis of gene expression regulated by transposable elements in human cancers. We adopted an analysis pipeline for screening methods to detect cancer- specific expression from expressed human sequences. We developed a database (TECESdb) for understanding the mechanism of cancer development in relation to transposable elements. A total of 999 genes fused with transposable elements were found to be cancer-related in our analysis of the EST database. According to GO (Gene Ontology) analysis, the majority of the 999 cancer-specific genes have functional association with gene receptor, DNA binding, and kinase activity. Our data could contribute greatly to our understanding of human cancers in relation to transposable elements. Key words: Transposable elements, Cancer, Fusion gene, Bioinformatics, EST also appeared in open-reading frames of functional INTRODUCTION human genes (Yulug et al., 1995; Makalowski et al., 1999; The human genome is estimated to be composed of 45% Nekrutenko and Li, 2001; Huh et al., 2006). transposable elements (International Human Genome The L1 5’UTR element is known to have an antisense Sequencing Consortium 2001). -

EWSR1 Gene EWS RNA Binding Protein 1
EWSR1 gene EWS RNA binding protein 1 Normal Function The EWSR1 gene provides instructions for making the EWS protein, whose function is not completely understood. The EWS protein has two regions that contribute to its function. One region, the transcriptional activation domain, allows the EWS protein to turn on (activate) the first step in the production of proteins from genes (transcription). The other region, the RNA-binding domain, allows the EWS protein to attach (bind) to the genetic blueprint for proteins called RNA. The EWS protein may be involved in piecing together this blueprint. Some studies suggest that the RNA-binding domain is able to block (inhibit) the activity of the transcriptional activation domain, and thus regulate the function of the EWS protein. Health Conditions Related to Genetic Changes Ewing sarcoma Mutations involving the EWSR1 gene can cause a type of cancerous tumor known as Ewing sarcoma. These tumors develop in bones or soft tissues, such as nerves and cartilage. There are several types of Ewing sarcoma, including Ewing sarcoma of bone, extraosseous Ewing sarcoma, peripheral primitive neuroectodermal tumor, and Askin tumor. The mutations that cause these tumors are acquired during a person's lifetime and are present only in the tumor cells. This type of genetic change, called a somatic mutation, is not inherited. The most common mutation that causes Ewing sarcoma is a rearrangement (translocation) of genetic material between chromosome 22 and chromosome 11. This translocation, written as t(11;22), fuses part of the EWSR1 gene on chromosome 22 with part of another gene on chromosome 11 called FLI1, creating an EWSR1/FLI1 fusion gene. -

Identification and Characterization of RET Fusions in Advanced Colorectal Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 30 Identification and characterization of RET fusions in advanced colorectal cancer Anne-France Le Rolle1,2,*, Samuel J. Klempner1,2,*, Christopher R. Garrett3, Tara Seery1,2, Eric M. Sanford4, Sohail Balasubramanian4, Jeffrey S. Ross4,5, Philip J. Stephens4, Vincent A. Miller4, Siraj M. Ali4 and Vi K. Chiu1,2 1 Division of Hematology/Oncology, Department of Medicine, University of California Irvine, Irvine, CA, USA 2 Chao Family Comprehensive Cancer Center, University of California Irvine, Orange, CA, USA 3 The Division of Cancer Medicine, Department of Gastrointestinal Medical Oncology, MD Anderson Cancer Center, Houston, TX, USA 4 Foundation Medicine Inc., Cambridge, MA, USA 5 Albany Medical College, Albany, NY, USA * These authors have contributed equally to this work Correspondence to: Vi K. Chiu, email: [email protected] Keywords: RET fusion kinase, RET kinase inhibitor, comprehensive genomic profiling, colorectal cancer Received: April 02, 2015 Accepted: May 12, 2015 Published: May 30, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT There is an unmet clinical need for molecularly directed therapies available for metastatic colorectal cancer. Comprehensive genomic profiling has the potential to identify actionable genomic alterations in colorectal cancer. Through comprehensive genomic profiling we prospectively identified 6 RET fusion kinases, including two novel fusions of CCDC6-RET and NCOA4-RET, in metastatic colorectal cancer (CRC) patients. RET fusion kinases represent a novel class of oncogenic driver in CRC and occurred at a 0.2% frequency without concurrent driver mutations, including KRAS, NRAS, BRAF, PIK3CA or other fusion tyrosine kinases. -

Birth of a Chimeric Primate Gene by Capture of the Transposase Gene
Birth of a chimeric primate gene by capture of the SEE COMMENTARY transposase gene from a mobile element Richard Cordaux*, Swalpa Udit†, Mark A. Batzer*, and Ce´ dric Feschotte†‡ *Department of Biological Sciences, Biological Computation and Visualization Center, Center for BioModular Multi-Scale Systems, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803; and †Department of Biology, University of Texas, Arlington, TX 76019 Edited by Susan R. Wessler, University of Georgia, Athens, GA, and approved March 27, 2006 (received for review February 10, 2006) The emergence of new genes and functions is of central impor- SETMAR transcript, which consists of these three exons, is tance to the evolution of species. The contribution of various types predicted to encode a protein of 671 amino acids and is of duplications to genetic innovation has been extensively inves- supported by 48 human cDNA clones from 18 different normal tigated. Less understood is the creation of new genes by recycling and͞or cancerous tissues (Table 1, which is published as sup- of coding material from selfish mobile genetic elements. To inves- porting information on the PNAS web site; refs. 14 and 15). tigate this process, we reconstructed the evolutionary history of These data suggest that the SETMAR protein is broadly ex- SETMAR, a new primate chimeric gene resulting from fusion of a pressed and has an important, yet unknown, function in human. SET histone methyltransferase gene to the transposase gene of a Recently, it was shown that the SET domain of the SETMAR mobile element. We show that the transposase gene was recruited protein exhibits histone methyltransferase activity (15), as do all as part of SETMAR 40–58 million years ago, after the insertion of known SET domains (16, 17). -

Selective Induction of Leukemia-Associated Fusion Genes by High-Dose Ionizing Radiation1
[CANCER RESEARCH 58. 421-425. February I. 1<W8| Selective Induction of Leukemia-associated Fusion Genes by High-Dose Ionizing Radiation1 Michael W. N. Deininger, Shikha Bose, Joanna Gora-Tybor, Xiu-Hua Yan, John M. Goldman, and Junia V. Melo2 Leukaemia Research Fumi Centre for Adult Leukaemia. Department of Haemah>li>/;\: Royal Postgraduale Medical School, Ducane Road, London W12 ONN, United Kingdom ABSTRACT event involves the acquisition of the genetic abnormality whose "success" in the production of a leukemic phenotype will depend on There is strong clinical and epidemiológica! evidence that ionizing its capacity to impart to the target cell a proliferative and/or survival radiation can cause leukemia by inducing DNA damage. This crucial advantage over its normal neighbors. In molecular terms, the gener initiation event is believed to be the result of random DNA breakage and misrepair, whereas the subsequent steps, promotion and progression, ation of a potentially successful reciprocal chromosomal translocation must rely on mechanisms of selective pressure to provide the expanding requires that: (a) at least two independent DNA DSBs occur, one in leukemic population with its proliferative/renewal advantage. To investi each chromosome partner; (b) the two breaks occur simultaneously, gate the susceptibility of human cells to external agents at the genetic i.e., within the same cell cycle, so that the two ends of one broken recombination stage of leukemogenesis, we subjected two hematopoietic chromosome are available to interact and be ligated (misrepaired) to cell lines, KG1 and III.6(1, to high doses of y-irradiation. The irradiation the respective complementary broken ends of the other chromosome; induced the formation of fusion genes characteristic of leukemia in both and (c) the recombination observes the polarity of the DNA molecule. -

DNA Transposons and the Evolution of Eukaryotic Genomes
ANRV329-GE41-15 ARI 12 October 2007 11:1 DNA Transposons and the Evolution of Eukaryotic Genomes Cedric´ Feschotte and Ellen J. Pritham Department of Biology, University of Texas, Arlington, Texas 76019; email: [email protected] Annu. Rev. Genet. 2007. 41:331–68 Key Words The Annual Review of Genetics is online at transposable elements, transposase, molecular domestication, http://genet.annualreviews.org chromosomal rearrangements This article’s doi: 10.1146/annurev.genet.40.110405.090448 Abstract Copyright c 2007 by Annual Reviews. Transposable elements are mobile genetic units that exhibit broad All rights reserved by Fordham University on 11/23/12. For personal use only. diversity in their structure and transposition mechanisms. Transpos- 0066-4197/07/1201-0331$20.00 able elements occupy a large fraction of many eukaryotic genomes and their movement and accumulation represent a major force shap- Annu. Rev. Genet. 2007.41:331-68. Downloaded from www.annualreviews.org ing the genes and genomes of almost all organisms. This review fo- cuses on DNA-mediated or class 2 transposons and emphasizes how this class of elements is distinguished from other types of mobile elements in terms of their structure, amplification dynamics, and genomic effect. We provide an up-to-date outlook on the diversity and taxonomic distribution of all major types of DNA transposons in eukaryotes, including Helitrons and Mavericks. We discuss some of the evolutionary forces that influence their maintenance and di- versification in various genomic environments. Finally, we highlight how the distinctive biological features of DNA transposons have contributed to shape genome architecture and led to the emergence of genetic innovations in different eukaryotic lineages.