Gene Expression Profiling of Transporters in the Solute Carrier and ATP-Binding Cassette Superfamilies in Human Eye Substructures
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
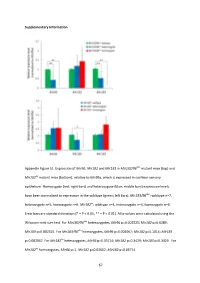
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

The Genetic Landscape of the Human Solute Carrier (SLC) Transporter Superfamily
Human Genetics (2019) 138:1359–1377 https://doi.org/10.1007/s00439-019-02081-x ORIGINAL INVESTIGATION The genetic landscape of the human solute carrier (SLC) transporter superfamily Lena Schaller1 · Volker M. Lauschke1 Received: 4 August 2019 / Accepted: 26 October 2019 / Published online: 2 November 2019 © The Author(s) 2019 Abstract The human solute carrier (SLC) superfamily of transporters is comprised of over 400 membrane-bound proteins, and plays essential roles in a multitude of physiological and pharmacological processes. In addition, perturbation of SLC transporter function underlies numerous human diseases, which renders SLC transporters attractive drug targets. Common genetic polymorphisms in SLC genes have been associated with inter-individual diferences in drug efcacy and toxicity. However, despite their tremendous clinical relevance, epidemiological data of these variants are mostly derived from heterogeneous cohorts of small sample size and the genetic SLC landscape beyond these common variants has not been comprehensively assessed. In this study, we analyzed Next-Generation Sequencing data from 141,456 individuals from seven major human populations to evaluate genetic variability, its functional consequences, and ethnogeographic patterns across the entire SLC superfamily of transporters. Importantly, of the 204,287 exonic single-nucleotide variants (SNVs) which we identifed, 99.8% were present in less than 1% of analyzed alleles. Comprehensive computational analyses using 13 partially orthogonal algorithms that predict the functional impact of genetic variations based on sequence information, evolutionary conserva- tion, structural considerations, and functional genomics data revealed that each individual genome harbors 29.7 variants with putative functional efects, of which rare variants account for 18%. Inter-ethnic variability was found to be extensive, and 83% of deleterious SLC variants were only identifed in a single population. -

Disease-Induced Modulation of Drug Transporters at the Blood–Brain Barrier Level
International Journal of Molecular Sciences Review Disease-Induced Modulation of Drug Transporters at the Blood–Brain Barrier Level Sweilem B. Al Rihani 1 , Lucy I. Darakjian 1, Malavika Deodhar 1 , Pamela Dow 1 , Jacques Turgeon 1,2 and Veronique Michaud 1,2,* 1 Tabula Rasa HealthCare, Precision Pharmacotherapy Research and Development Institute, Orlando, FL 32827, USA; [email protected] (S.B.A.R.); [email protected] (L.I.D.); [email protected] (M.D.); [email protected] (P.D.); [email protected] (J.T.) 2 Faculty of Pharmacy, Université de Montréal, Montreal, QC H3C 3J7, Canada * Correspondence: [email protected]; Tel.: +1-856-938-8697 Abstract: The blood–brain barrier (BBB) is a highly selective and restrictive semipermeable network of cells and blood vessel constituents. All components of the neurovascular unit give to the BBB its crucial and protective function, i.e., to regulate homeostasis in the central nervous system (CNS) by removing substances from the endothelial compartment and supplying the brain with nutrients and other endogenous compounds. Many transporters have been identified that play a role in maintaining BBB integrity and homeostasis. As such, the restrictive nature of the BBB provides an obstacle for drug delivery to the CNS. Nevertheless, according to their physicochemical or pharmacological properties, drugs may reach the CNS by passive diffusion or be subjected to putative influx and/or efflux through BBB membrane transporters, allowing or limiting their distribution to the CNS. Drug transporters functionally expressed on various compartments of the BBB involve numerous proteins from either the ATP-binding cassette (ABC) or the solute carrier (SLC) superfamilies. -

Nephritis Responses in Murine and Human Lupus Analysis Defines
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 24 of which you can access for free at: 2012; 189:988-1001; Prepublished online 20 June from submission to initial decision 4 weeks from acceptance to publication Celine C. Berthier, Ramalingam Bethunaickan, Tania Gonzalez-Rivera, Viji Nair, Meera Ramanujam, Weijia Zhang, Erwin P. Bottinger, Stephan Segerer, Maja Lindenmeyer, Clemens D. Cohen, Anne Davidson and Matthias Kretzler 2012; doi: 10.4049/jimmunol.1103031 http://www.jimmunol.org/content/189/2/988 Cross-Species Transcriptional Network Analysis Defines Shared Inflammatory Responses in Murine and Human Lupus Nephritis J Immunol cites 60 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/189/2/988.full#ref-list-1 http://www.jimmunol.org/content/suppl/2012/06/20/jimmunol.110303 1.DC1 This article Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. The Journal of Immunology Cross-Species Transcriptional Network Analysis Defines Shared Inflammatory Responses in Murine and Human Lupus Nephritis Celine C. -

The Genetics of Bipolar Disorder
Molecular Psychiatry (2008) 13, 742–771 & 2008 Nature Publishing Group All rights reserved 1359-4184/08 $30.00 www.nature.com/mp FEATURE REVIEW The genetics of bipolar disorder: genome ‘hot regions,’ genes, new potential candidates and future directions A Serretti and L Mandelli Institute of Psychiatry, University of Bologna, Bologna, Italy Bipolar disorder (BP) is a complex disorder caused by a number of liability genes interacting with the environment. In recent years, a large number of linkage and association studies have been conducted producing an extremely large number of findings often not replicated or partially replicated. Further, results from linkage and association studies are not always easily comparable. Unfortunately, at present a comprehensive coverage of available evidence is still lacking. In the present paper, we summarized results obtained from both linkage and association studies in BP. Further, we indicated new potential interesting genes, located in genome ‘hot regions’ for BP and being expressed in the brain. We reviewed published studies on the subject till December 2007. We precisely localized regions where positive linkage has been found, by the NCBI Map viewer (http://www.ncbi.nlm.nih.gov/mapview/); further, we identified genes located in interesting areas and expressed in the brain, by the Entrez gene, Unigene databases (http://www.ncbi.nlm.nih.gov/entrez/) and Human Protein Reference Database (http://www.hprd.org); these genes could be of interest in future investigations. The review of association studies gave interesting results, as a number of genes seem to be definitively involved in BP, such as SLC6A4, TPH2, DRD4, SLC6A3, DAOA, DTNBP1, NRG1, DISC1 and BDNF. -

Roles for the Uptake 2 Transporter OCT3 in Regulation Of
Marquette University e-Publications@Marquette Biomedical Sciences Faculty Research and Publications Biomedical Sciences, Department of 2-2019 Roles for the Uptake2 Transporter OCT3 in Regulation of Dopaminergic Neurotransmission and Behavior Paul J. Gasser Marquette University, [email protected] Follow this and additional works at: https://epublications.marquette.edu/biomedsci_fac Part of the Neurosciences Commons Recommended Citation Gasser, Paul J., "Roles for the Uptake2 Transporter OCT3 in Regulation of Dopaminergic Neurotransmission and Behavior" (2019). Biomedical Sciences Faculty Research and Publications. 191. https://epublications.marquette.edu/biomedsci_fac/191 Marquette University e-Publications@Marquette Biomedical Sciences Faculty Research and Publications/College of Health Sciences This paper is NOT THE PUBLISHED VERSION; but the author’s final, peer-reviewed manuscript. The published version may be accessed by following the link in the citation below. Neurochemistry International, Vol. 123, (February 2019): 46-49. DOI. This article is © Elsevier and permission has been granted for this version to appear in e-Publications@Marquette. Elsevier does not grant permission for this article to be further copied/distributed or hosted elsewhere without the express permission from Elsevier. Roles for the Uptake2 Transporter OCT3 in Regulation of Dopaminergic Neurotransmission and Behavior Paul J. Gasser Department of Biomedical Sciences, Marquette University, Milwaukee, WI Abstract Transporter-mediated uptake determines the -

SLC12A6 Gene Solute Carrier Family 12 Member 6
SLC12A6 gene solute carrier family 12 member 6 Normal Function The SLC12A6 gene provides instructions for making a protein called a K-Cl cotransporter. This protein is involved in moving charged atoms (ions) of potassium (K) and chlorine (Cl) across the cell membrane. The positively charged potassium ions and negatively charged chlorine ions are moved together (cotransported), so that the charges inside and outside the cell membrane are unchanged (electroneutral). Electroneutral cotransport of ions across cell membranes is involved in many functions of the body. While the specific function of the K-Cl cotransporter produced from the SLC12A6 gene is unknown, it seems to be critical for the development and maintenance of nerve tissue. It may be involved in regulating the amounts of potassium, chlorine, or water in cells and intercellular spaces. The K-Cl cotransporter protein may also help regulate the activity of other proteins that are sensitive to ion concentrations. Health Conditions Related to Genetic Changes Andermann syndrome At least six SLC12A6 gene mutations have been identified in people with Andermann syndrome. Almost all affected individuals of French-Canadian descent have the same mutation in both copies of the SLC12A6 gene, in which the DNA building block ( nucleotide) guanine is deleted at position 2436 (written as 2436delG). This mutation is common in the populations of the Saguenay-Lac-St.-Jean and Charlevoix regions of northeastern Quebec. Most SLC12A6 gene mutations that cause Andermann syndrome result in a K-Cl cotransporter protein that is shortened and nonfunctional. The lack of functional protein produced from the SLC12A6 gene is believed to interfere with the development of the corpus callosum and maintenance of the nerves that transmit signals needed for movement and sensation, resulting in the signs and symptoms of Andermann syndrome. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

SLC22A3 Is Associated with Lipoprotein (A) Concentration And
Clinical Biochemistry 66 (2019) 44–48 Contents lists available at ScienceDirect Clinical Biochemistry journal homepage: www.elsevier.com/locate/clinbiochem SLC22A3 is associated with lipoprotein (a) concentration and cardiovascular disease in familial hypercholesterolemia T ⁎ Martine Paquettea, Sophie Bernarda,b, Alexis Baassa,c,d, a Lipids, Nutrition and Cardiovascular Prevention Clinic, Montreal Clinical Research Institute, Québec, Canada b Department of Medicine, Division of Endocrinology, Université de Montreal, Québec, Canada c Department of Medicine, Division of Experimental Medicine, McGill University, Québec, Canada d Department of Medicine, Division of Medical Biochemistry, McGill University, Québec, Canada ARTICLE INFO ABSTRACT Keywords: Background and aims: Several clinical and genetic factors have been shown to modulate the cardiovascular risk Familial hypercholesterolemia in subjects affected by familial hypercholesterolemia (FH). Genome wide association studies (GWAS) in the Cardiovascular disease general population have identified several single nucleotide polymorphisms (SNPs) significantly associated with SLC22A3 the risk of cardiovascular disease (CVD). This include the rs2048327 variant in the SLC22A3 gene. However, the Lipoprotein (a) effect of this SNP in FH subjects is unknown. The objectives of this study are to investigate the association Risk stratification between rs2048327 and the prevalence of CVD as well as with the concentration of lipoprotein (a) (Lp (a)), in a rs2048327 cohort of genetically-confirmed heterozygous FH patients. Methods: An enzyme-linked immunoassay kit was used to assess the Lp (a) concentration, whereas an exome chip genotyping method was used to impute the rs2048327 genotype. Results: The cohort comprised 287 non-carriers (TT), 305 heterozygous carriers (TC) and 76 homozygous car- riers of the rs2048327 variant. In a model corrected for traditional cardiovascular risk factors, rs2048327 was significantly associated with Lp (a) level (median value of 12, 16 and 29 mg/dL in TT, TC and CC carriers, respectively, p < .0001). -

Genetic Background of Ataxia in Children Younger Than 5 Years in Finland E444
Volume 6, Number 4, August 2020 Neurology.org/NG A peer-reviewed clinical and translational neurology open access journal ARTICLE Genetic background of ataxia in children younger than 5 years in Finland e444 ARTICLE Cerebral arteriopathy associated with heterozygous variants in the casitas B-lineage lymphoma gene e448 ARTICLE Somatic SLC35A2 mosaicism correlates with clinical fi ndings in epilepsy brain tissuee460 ARTICLE Synonymous variants associated with Alzheimer disease in multiplex families e450 Academy Officers Neurology® is a registered trademark of the American Academy of Neurology (registration valid in the United States). James C. Stevens, MD, FAAN, President Neurology® Genetics (eISSN 2376-7839) is an open access journal published Orly Avitzur, MD, MBA, FAAN, President Elect online for the American Academy of Neurology, 201 Chicago Avenue, Ann H. Tilton, MD, FAAN, Vice President Minneapolis, MN 55415, by Wolters Kluwer Health, Inc. at 14700 Citicorp Drive, Bldg. 3, Hagerstown, MD 21742. Business offices are located at Two Carlayne E. Jackson, MD, FAAN, Secretary Commerce Square, 2001 Market Street, Philadelphia, PA 19103. Production offices are located at 351 West Camden Street, Baltimore, MD 21201-2436. Janis M. Miyasaki, MD, MEd, FRCPC, FAAN, Treasurer © 2020 American Academy of Neurology. Ralph L. Sacco, MD, MS, FAAN, Past President Neurology® Genetics is an official journal of the American Academy of Neurology. Journal website: Neurology.org/ng, AAN website: AAN.com CEO, American Academy of Neurology Copyright and Permission Information: Please go to the journal website (www.neurology.org/ng) and click the Permissions tab for the relevant Mary E. Post, MBA, CAE article. Alternatively, send an email to [email protected]. -

Pet-1 Controls Tetrahydrobiopterin Pathway and Slc22a3 Transporter Genes in Serotonin Neurons Steven C Wyler, Lauren J Donovan, Mia Yeager, and Evan Deneris ACS Chem
Subscriber access provided by CASE WESTERN RESERVE UNIV Article Pet-1 controls tetrahydrobiopterin pathway and Slc22a3 transporter genes in serotonin neurons Steven C Wyler, Lauren J Donovan, Mia Yeager, and Evan Deneris ACS Chem. Neurosci., Just Accepted Manuscript • Publication Date (Web): 02 Feb 2015 Downloaded from http://pubs.acs.org on February 5, 2015 Just Accepted “Just Accepted” manuscripts have been peer-reviewed and accepted for publication. They are posted online prior to technical editing, formatting for publication and author proofing. The American Chemical Society provides “Just Accepted” as a free service to the research community to expedite the dissemination of scientific material as soon as possible after acceptance. “Just Accepted” manuscripts appear in full in PDF format accompanied by an HTML abstract. “Just Accepted” manuscripts have been fully peer reviewed, but should not be considered the official version of record. They are accessible to all readers and citable by the Digital Object Identifier (DOI®). “Just Accepted” is an optional service offered to authors. Therefore, the “Just Accepted” Web site may not include all articles that will be published in the journal. After a manuscript is technically edited and formatted, it will be removed from the “Just Accepted” Web site and published as an ASAP article. Note that technical editing may introduce minor changes to the manuscript text and/or graphics which could affect content, and all legal disclaimers and ethical guidelines that apply to the journal pertain. ACS cannot be held responsible for errors or consequences arising from the use of information contained in these “Just Accepted” manuscripts. ACS Chemical Neuroscience is published by the American Chemical Society.