Identification of Common Adulterants in Walnut Beverage Based on Plant DNA Barcode Technology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
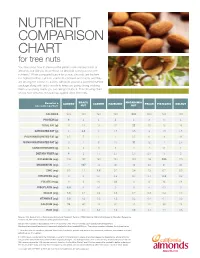
Nutrient Comparison Chart
NUTRIENT COMPARISON CHART for tree nuts You may know how to measure the perfect one-ounce portion of almonds, but did you know those 23 almonds come packed with nutrients? When compared ounce for ounce, almonds are the tree nut highest in fiber, calcium, vitamin E, riboflavin and niacin, and they are among the lowest in calories. Almonds provide a powerful nutrient package along with tasty crunch to keep you going strong, making them a satisfying snack you can feel good about. The following chart shows how almonds measure up against other tree nuts. BRAZIL MACADAMIA Based on a ALMOND CASHEW HAZELNUT PECAN PISTACHIO WALNUT one-ounce portion1 NUT NUT CALORIES 1602 190 160 180 200 200 160 190 PROTEIN (g) 6 4 4 4 2 3 6 4 TOTAL FAT (g) 14 19 13 17 22 20 13 19 SATURATED FAT (g) 1 4.5 3 1.5 3.5 2 1.5 1.5 POLYUNSATURATED FAT (g) 3.5 7 2 2 0.5 6 4 13 MONOUNSATURATED FAT (g) 9 7 8 13 17 12 7 2.5 CARBOHYDRATES (g) 6 3 9 5 4 4 8 4 DIETARY FIBER (g) 4 2 1.5 2.5 2.5 2.5 3 2 POTASSIUM (mg) 208 187 160 193 103 116 285 125 MAGNESIUM (mg) 77 107 74 46 33 34 31 45 ZINC (mg) 0.9 1.2 1.6 0.7 0.4 1.3 0.7 0.9 VITAMIN B6 (mg) 0 0 0.1 0.2 0.1 0.1 0.3 0.2 FOLATE (mcg) 12 6 20 32 3 6 14 28 RIBOFLAVIN (mg) 0.3 0 0.1 0 0 0 0.1 0 NIACIN (mg) 1.0 0.1 0.4 0.5 0.7 0.3 0.4 0.3 VITAMIN E (mg) 7.3 1.6 0.3 4.3 0.2 0.4 0.7 0.2 CALCIUM (mg) 76 45 13 32 20 20 30 28 IRON (mg) 1.1 0.7 1.7 1.3 0.8 0.7 1.1 0.8 Source: U.S. -

Pistachio and Walnut Development Project TCP/KYR/3203 (D)
State Agency of Forestry and Environmental Protection, Kyrgyz Republic Pistachio and Walnut Development Project TCP/KYR/3203 (D) Hafiz Muminjanov Plant Production & Protection Officer Food and Agriculture Organization of the United Nations Sub-regional Office for Central Asia (FAOSEC) Agriculture in Kyrgyz Republic • Cereals - 1.5 mln MT, • including wheat 0.7 mln. MT • Potato - 1. 3 mln. MT • Vegetables - 0.8 mln. MT • Fruits -200 000 MT • Grapes - 10 000 MT 1 Why pistachio and walnut? Total Plantation Production, Yield, Area, ha Area, ha MT kg/ha Pistachio 57 000 12000 1 000 15-20 Walnut 47 000 - 900-1 900 20-40 Agriculture growth vs Overall Growth Kyrgyz Republic - Ag. Growth Vs. Overall Growth 160 Total GDP (1990=100) 140 AgGDP (1990=100) Total GDP (2002=100) AgGDP (2002=100) 120 100 AgGDP 80 (1990=100) Total GDP 60 (1990=100) 40 2 Wheat Grain Trade Balance ('000 tons) 1,200 1,000 800 600 Import 400 Domestic Production 200 - 2003 2004 2005 2006 2007 Trade Balance Values Dry Nuts Vs. Agriculture (2001 = 100) 600 Dry Nuts 400 200 0 2001 2002 2003 2004 2005 2006 - 200 - 400 Agriculture - 600 3 Why pistachio and walnut? • Central Asia – centre of origin & diversity of nuts • Large area of forest plantations • Suitable climatic conditions (dry summer) • Suitable socio-economic environment • Organic forests 4 Why pistachio and walnut? • Absence of market orientation in the past • Current varieties not valued for international market • No management done (cutting, pruning, etc.) • Density is high, pistachio male/female ratio not regulated • Yield is low • History of quality produce • International standards (USA, France, Chile, Australia) • Demand increase • Potential for export • Increasing farmers income & Livelihood improvement 5 Why pistachio and walnut? Development of the nut fruit sector has a high potential in improving livelihood and food security due the high international demand… Past and Related Work • Tien Shan Ecosystem Development Project (IFAD; GEF; WB-Bio Carbon Fund; Japanese Government). -
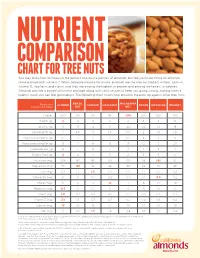
Chart for Tree Nuts
NUTRIENT COMPARISON CHART FOR TREE NUTS You may know how to measure the perfect one-ounce portion of almonds, but did you know those 23 almonds come packed with nutrients? When compared ounce for ounce, almonds are the tree nut highest in fiber, calcium, vitamin E, riboflavin and niacin, and they are among the highest in protein and among the lowest in calories. Almonds provide a powerful nutrient package along with tasty crunch to keep you going strong, making them a healthy snack you can feel good about. The following chart shows how almonds measure up against other tree nuts. BRAZIL MACADAMIA Based on a ALMOND CASHEW HAZELNUT PECAN PISTACHIO WALNUT one-ounce portion1 NUT NUT Calories 1602 190 160 180 200 200 160 190 Protein (g) 6 4 4 4 2 3 6 4 Total Fat (g) 14 19 13 17 22 20 13 18 Saturated Fat (g) 1 4.5 3 1.5 3.5 2 1.5 1.5 Polyunsaturated Fat (g) 3.5 7 2 2 0.5 6 4 13 Monounsaturated Fat (g) 9 7 8 13 17 12 7 2.5 Carbohydrates (g) 6 3 9 5 4 4 8 4 Dietary Fiber (g) 4 2 1 3 2 3 3 2 Potassium (mg) 208 187 160 193 103 116 285 125 Magnesium (mg) 77 107 74 46 33 34 31 45 Zinc (mg) 0.9 1.2 1.6 0.7 0.4 1.3 0.7 0.9 Vitamin B6 (mg) 0 0 0.1 0.2 0.1 0.1 0.3 0.2 Folate (mcg) 12 6 20 32 3 6 14 28 Riboflavin (mg) 0.3 0 0.1 0 0 0 0.1 0 Niacin (mg) 1.0 0.1 0.4 0.5 0.7 0.3 0.4 0.3 Vitamin E (mg) 7.3 1.6 0.3 4.3 0.2 0.4 0.6 0.2 Calcium (mg) 76 45 13 32 20 20 30 28 Iron (mg) 1.1 0.7 1.7 1.3 0.8 0.7 1.1 0.8 Source: U.S. -

Tree Nut Allergen Component Testing Brochure
Walnut, cashew, and Brazil nut allergen component testing Jug r1 Walnut, Jug cashew, and r3 Brazil nut allergens by Ana o3 the numbers Ber e1 Detect sensitizations to the whole nut to create personalized management plans for your patients. Allergen component testing Measurement of specific IgE by blood test that provides objective assessment of sensitization to the whole nut protein is the first step in discovering the likelihood of a systemic reaction and the necessary precautions that may be prescribed. Walnut TC 3489 One of the most common causes WHOLE of allergic reactions to tree nuts.1,2 Walnut ALLERGEN • Estimated prevalence of walnut allergy in the general population is up to 0.7%.2 • Potentially life-threatening, increasing in prevalence and rarely outgrown.2,3 Associated with systemic Jug reactions2 r1 • Storage protein (2s albumin) 3,4 4 ALLERGEN • Heat and digestion stable COMPONENTS • Highly abundant in walnut Associated with local and Jug systemic reactions2 r3 • Lipid transfer protein (LTP)1,4 • Heat and digestion stable Knowing which Positive whole walnut with negative protein your patient is Jug r1 and Jug r3 results may be explained by sensitization to6: sensitized to can help • Other walnut storage proteins you develop a • Pollen proteins like profilin or PR-10 proteins management plan. • CCD (cross-reacting carbohydrate determinants) Cashew nut Brazil nut TC 2608 TC 2818 Allergy on the rise with increased Hidden allergen often found Cashew consumption in snack foods, Asian Brazil in cookies, insect repellent foods, baked goods, nut butters and and beauty products.12 pestos.8,9 • Extensive cross-reactivity within • Sensitized patients have a risk of experiencing the family can be expected.13 severe allergic reactions; the risk has been • Generally persists and is potentially reported to be even higher than for peanut life-threatening.14 allergic patients (74% vs. -

Black Walnut Juglans Nigra
black walnut Juglans nigra Kingdom: Plantae FEATURES Division: Magnoliophyta The deciduous black walnut tree may grow to a Class: Magnoliopsida height of 150 feet and a diameter of five feet. The Order: Fagales trunk is straight, and the crown is rounded. The bark is thick, black and deeply furrowed. The pith in the Family: Juglandaceae twigs is chambered, that is, divided by partitions. ILLINOIS STATUS The bud is rounded at the tip, pale brown and hairy. The pinnately compound leaves have 15 to 23 common, native leaflets and are arranged alternately on the stem. © Guy Sternberg Each lance-shaped leaflet may be up to three and one-half inches long and one and one-half inches wide. The leaflet is toothed along the edges, yellow- green and smooth above and paler and hairy below. Leaves turn yellow in the fall. Male and female flowers are separate but located on the same tree. The male (staminate) flowers are arranged in yellow- green, hairy catkins, while the female (pistillate) flowers are in small spikes. Neither type of flower has petals. The spherical fruits are arranged in groups of one or two. Each green or yellow-green walnut may be up to two inches in diameter. The husk on the fruit is thick, while the nut is very hard, oval, dark brown and deeply ridged. The seed is sweet to the taste. tree in summer BEHAVIORS The black walnut may be found statewide in Illinois. ILLINOIS RANGE This tree grows in rich woodlands. The black walnut flowers in April and May when the leaves are partly grown. -

Identification of Butternuts and Butternut Hybrids
Purdue University Purdue extension FNR-420-W & Natural Re ry sou Forestry and Natural Resources st rc re e o s F Identification of Butternuts and Butternut Hybrids Lenny Farlee1,3, Keith Woeste1, Michael Ostry2, James McKenna1 and Sally Weeks3 1 USDA Forest Service Hardwood Tree Improvement and Regeneration Center, Purdue University, 715 W. State Street, West Lafayette, IN, 47907 PURDUE UNIVERSITY 2 USDA Forest Service Northern Research Station, 1561 Lindig Ave. St. Paul, MN 55108 3 Department of Forestry and Natural Resources, Purdue University, 715 W. State Street, West Lafayette, IN, 47907 Introduction Butternut (Juglans cinerea), also known as white walnut, is a native hardwood related to black walnut (Juglans nigra) and other members of the walnut family. Butternut is a medium-sized tree with alternate, pinnately compound leaves that bears large, sharply ridged and corrugated, elongated, cylindrical nuts born inside sticky green hulls that earned it the nickname lemon-nut (Rink, 1990). The nuts are a preferred food of squirrels and other wildlife. Butternuts were collected and eaten by Native Americans (Waugh, 1916; Hamel and Chiltoskey, 1975) and early settlers, who also valued butternut for its workable, medium brown-colored wood (Kellogg, 1919), and as a source of medicine (Johnson, 1884), dyes (Hamel and Chiltoskey, 1975), and sap sugar. Butternut’s native range extends over the entire north- eastern quarter of the United States, including many states immediately west of the Mississippi River, and into Canada. Butternut is more cold-tolerant than black walnut, and it grows as far north as the Upper Peninsula of Michigan, New Brunswick, southern Quebec, and Figure 1. -

Harvesting and Storing Your Home Orchard's Nut Crop
PUBLICATION 8005 Harvesting and Storing Your Home Orchard’s Nut Crop: Almonds, Walnuts, Pecans, Pistachios, and Chestnuts UNIVERSITY OF CALIFORNIA ED PERRY, University of California Cooperative Extension Farm Advisor, Division of Agriculture Stanislaus County; G. STEVEN SIBBETT, University of California Cooperative Extension and Natural Resources Farm Advisor, Tulare County. http://danrcs.ucdavis.edu he quality and quantity of your home orchard’s nut crop depend on the harvest- Ting, processing, and storage techniques that you use. This publication offers advice for handling almonds, chestnuts, pecans, pistachios, and walnuts. GENERAL TIPS When to Harvest Harvest all types of nuts as soon as they are ready. Late harvesting reduces crop volume, lowers nut quality, and shortens storage life. In California, harvest timing varies somewhat with location. The earliest harvest generally occurs in the southern San Joaquin Valley, followed by the central San Joaquin Valley, the Sacramento Valley, and finally the coastal valleys. An interval of about seven days separates the begin- ning dates for harvest in each succeeding district. Harvesting To harvest nuts (except chestnuts), knock or shake them from their trees with long wooden, fiberglass, or plastic (PVC) poles. You can knock almonds from the trees using rubber mallets or poles that are available from farm supply stores. CAUTION: When harvesting nuts with a pole, use extreme caution. Any contact between the pole and power lines will result in serious injury or death. Do not use aluminum or other metal poles to harvest nuts: these are especially hazardous around power lines. Loosened nuts often follow the pole to the ground, striking the person holding the pole. -

Walnuts and Pecans - Nebraska Style
Walnuts and Pecans - Nebraska Style Based on 35 years of research by the Nebraska Nut Growers Association and the University of Nebraska- Lincoln, numerous landowners have planted walnut and pecan trees for commercial nut production in southeast Nebraska and adjacent states. These trees have matured and began to produce crops of nuts, marketing is now the issue. In 2005, members of Nebraska Nut Growers Association (NeNGA), recognizing the marketing problem, formed Heartland Nuts ‘N More (HN’NM) a cooperatively-owned business to assist nut growers in selling their production of nuts & nut products. Initially, 32 members made financial commitments to form the cooperative. HN’NM now boasts 46 members from four states: Nebraska, Iowa, Kansas, and Missouri. Their financial commitment provided opportunities to secure grants from a variety of sources. “If we hadn’t received those grants, we probably wouldn’t have started up at all.” said HN’NM spokesman Larry Martin. Grant funds were used to purchase a variety of equipment. A mechanized “tree shaker” loosens nuts from trees, allowing the nuts to be collected on tarps or plastic. HN’NM also has a mechanized “nut harvester” to collect nuts off the ground. Collected nuts are brought to the coop’s headquarters building in Valparaiso, NE, which also serves as the nut processing plant where a series of machines crack, sort, and package the nut meats. The nuts are stored in a cooler until sold. The highest quality black walnuts and pecans are sold in several local stores in Omaha and Lincoln including, Boiler Room, Whole Foods, Open Harvest, Ideal Foods, and Grow Nebraska. -

Brazil Nut: Nutritional Benefits from a Unique Combination of Antioxidants
Journal of International Society for Food Bioactives Nutraceuticals and Functional Foods Review J. Food Bioact. 2020;9:36–39 Brazil nut: nutritional benefits from a unique combination of antioxidants Alan Giovanini de Oliveira Sartoria*, Marisa A.B. Regitano-d’Arcea and Leif H. Skibstedb aDepartment of Agri-food Industry, Food and Nutrition, Luiz de Queiroz College of Agriculture, University of São Paulo, Piracicaba, SP, Brazil bDepartment of Food Science, Faculty of Science, University of Copenhagen, Frederiksberg, Denmark *Corresponding author: Alan Giovanini de Oliveira Sartori, Department of Agri-food Industry, Food and Nutrition, Luiz de Queiroz Col- lege of Agriculture, University of São Paulo, Piracicaba, SP, Brazil. E-mail: [email protected] DOI: 10.31665/JFB.2020.9216 Received: March 27, 2020; Revised received & accepted: March 31, 2020 Citation: de Oliveira Sartori, A.G, Regitano-d’Arce, M.A.B., and Skibsted, L.H. (2020). Brazil nut: nutritional benefits from a unique com- bination of antioxidants. J. Food Bioact. 9: 36–39. Abstract Brazil nut is the seed of the Bertholletia excelsa tree, which grows naturally in the rainforests of South America. The production chain is environmentally sustainable as seeds are collected from the hard-shelled indehiscent fruits as they fall on the forest floor, without the need of deforestation. Brazil nut is among the most popular and commercially important tree nuts, and a reduced risk of coronary heart disease among other health benefits have been related to its consumption. A high content of both soluble and bound phenolic antioxidants, especially in the brown skin of the kernel, partly explains this positive health effect together with a high content of oleic acid and linoleic acid. -

Avoiding Peanut, Tree Nuts, Egg, Corn, and Wheat Ingredients Common Food Allergens May Be Listed Many Different Ways on Food Labels and Can Be Hidden in Common Foods
Food Allergen Graph: Avoiding Peanut, Tree Nuts, Egg, Corn, and Wheat Ingredients Common food allergens may be listed many different ways on food labels and can be hidden in common foods. Below you will find different labels for common allergens. Avoiding Peanuts: Avoiding Tree Nuts: Avoiding Egg: Avoiding Corn: Avoiding Wheat: Artificial nuts Almond Albumin / albumen Corn - meal, flakes, Bread Crumbs Beer nuts Artificial nuts Egg (dried, powdered, syrup, solids, flour, Bulgur Cold pressed, expeller Brazil nut solids, white, yolk) niblets, kernel, Cereal extract Flour: pressed or extruded peanut Beechnut Eggnog alcohol, on the cob, Club Wheat all-purpose Butternut oil Globulin / Ovoglobulin starch, bread,muffins Conscous bread Goobers Cashew Fat subtitutes sugar/sweetener, oil, Cracker meal cake Durum Ground nuts Chestnut Livetin Caramel corn / flavoring durum Einkorn Mandelonas (peanuts soaked Chinquapin nut Lysozyme Citric acid (may be corn enriched Emmer in almond flavoring) Coconut (really is a fruit not a based) graham Mayonnaise Farina Mixed nuts tree nut, but classified as a Grits high gluten Meringue (meringue Hydrolyzed Monkey nuts nut on some charts) high protein powder) Hominy wheat protein Nut meat Filbert / hazelnut instant Ovalbumin Maize Kamut Gianduja -a chocolate-nut mix pastry Nut pieces Ovomucin / Ovomucoid / Malto / Dextrose / Dextrate Matzoh Peanut butter Ginkgo nut Modified cornstarch self-rising Ovotransferrin Matzoh meal steel ground Peanut flour Hickory nut Polenta Simplesse Pasta stone ground Peanut protein hydrolysate -

Cakes Jumbo Cupcakes Cheesecakes Pies Pastries
CAKES PIES Prices as ring 2016 Small (6-8 servings) 21.95 Small Cake 10-12 servings Large (16-20 servings) 37.95 Large Cake 16-20 servings *Half Sheet Cake 50-60 servings* $75-80 Coconut Cream Add Edible Image to Any Cake - $10 Chocolate Banana Cream Banana Cream Keylime *Chocolate Mousse Cake - sm 39.95 lg 59.95 Peanut Butter Chiffon cake layers, chocolate mousse filling, Belgian sprinkles Chocolate Cream *Coconut Cream Cake - sm 39.95 lg 59.95 Fresh Fruit & Custard White cake layers, coconut cream filling, whipped cream exterior Apple Pie (small only) 18.95 *Raspberry White Chocolate Mousse Cake - sm 39.95 lg 59.95 Blueberry Crumb Pie (small only) 18.95 White cake layers, white chocolate mousse and raspberry filling, decorated with raspberry mousse & white chocolate curls Death by Chocolate - sm 39.95 lg 59.95 CHEESECAKES Flourless chocolate cake layers, kettle cooked fudge, Belgian sprinkles Small (10-12 servings) 38.95 Million Dollar Cake - sm 39.95 lg 59.95 Large (16-20 servings) 58.95 Three decadent layers; authentic chocolate mousse, NY style cheesecake, flourless chocolate cake Raspberry White Chocolate Cheesecake German Chocolate Cake - sm 39.95 lg 59.95 Chocolate Cheesecake Classic cake & filling wrapped in kettle cooked fudge & pecans Turtle Cheesecake *Oreo® Cookies & Cream Cake - sm 39.95 lg 59.95 Variety Cheesecake White & devils food cake layers, oreo® cookie mousse *Keylime Cheesecake* *Carrot Cake - sm 39.95 lg 59.95 *Butterfinger Cheesecake* Carrot cake layers, cream cheese frosting, walnuts *Red Velvet Cheesecake* *Red -

Sensational Summer Berries It Is Summer Time and That Means Fresh Eating
April 2012 Newsletter July Sensational Summer Berries It is summer time and that means fresh eating. One way to enjoy more nutritious and delicious summer foods is to include more sensational summer berries! Berries encompass tantalizing tastes for everyone to enjoy from tart blackberries to sweet blueberries; there is a berry for everyone. These nutritional powerhouses deserve a place in everyone’s diet because they can aid in disease prevention. Blueberries are a great choice because they are a sweet summertime treat that are a good source of fiber and vitamin C and loaded with phytonutrients which can help ward off diseases like heart disease and cancer. Select firm, plump blueberries with uniform size. Add frozen blueberries to a smoothie or top your favorite yogurt with fresh blueberries for a quick snack. Blackberries are another sensational summer berry to include in your daily fruit consumption. They are good source of fiber, vitamin C and vitamin K. One cup of blackberries has 7 grams of dietary fiber which helps support a healthy gut. Blackberries are also full of phytonutrients like anthocyanin, which gives the berry its dark color. Raspberries are high in fiber and vitamin C and can come in several colors like red, black and golden. Choose dry, plump raspberries and avoid washing until you are ready to eat them. Need a fun way to get more raspberries in your diet? Make a fruit salsa with raspberries to top grilled fish or make raspberries the sweet topping on your light summer salad. Strawberries are one of the top favored fruits among children and adults.