Profiling Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

IRAK4 Gene Interleukin 1 Receptor Associated Kinase 4
IRAK4 gene interleukin 1 receptor associated kinase 4 Normal Function The IRAK4 gene provides instructions for making a protein that plays an important role in innate immunity, which is the body's early, nonspecific response to foreign invaders ( pathogens). The IRAK-4 protein is part of a signaling pathway that is involved in early recognition of pathogens and the initiation of inflammation to fight infection. In particular, the IRAK-4 protein relays signals from proteins called Toll-like receptors and IL-1 receptor-related proteins. As one of the first lines of defense against infection, Toll-like receptors recognize patterns that are common to many pathogens, rather than recognizing specific pathogens, and stimulate a quick immune response. The IL-1 receptor and related proteins recognize immune system proteins called cytokines that signal the need for an immune response. The resulting signaling pathway triggers inflammation, a nonspecific immune response that helps fight infection. Health Conditions Related to Genetic Changes IRAK-4 deficiency At least 20 mutations in the IRAK4 gene have been identified in people with IRAK-4 deficiency, an immune system disorder that leads to recurrent invasive bacterial infections. These gene mutations lead to an abnormally short, nonfunctional IRAK-4 protein or no protein at all. The loss of functional IRAK-4 protein blocks the initiation of inflammation in response to pathogens or cytokines that would normally help fight the infections. Because the early immune response is insufficient, bacterial -

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Clustering of the Structures of Protein Kinase Activation Loops
bioRxiv preprint doi: https://doi.org/10.1101/395723; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Clustering of the structures of protein kinase activation loops: A new nomenclature for active and inactive kinase structures Vivek Modi Roland L. Dunbrack, Jr.* Institute for Cancer Research Fox Chase Cancer Center Philadelphia PA 19111 *[email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/395723; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract TarGeting protein kinases is an important strateGy for intervention in cancer. Inhibitors are directed at the conserved active conformation or a variety of inactive conformations. While attempts have been made to classify these conformations, a structurally rigorous cataloGue of states has not been achieved. The kinase activation loop is crucial for catalysis and beGins with the conserved DFGmotif (Asp-Phe-Gly). This motif is observed in two major classes of conformations, DFGin - an ensemble of active and inactive conformations where the Phe residue is in contact with the C-helix of the N-terminal lobe, and DFGout - an inactive form where Phe occupies the ATP site exposinG the C-helix pocket. -

Genetic Mutations and Mechanisms in Dilated Cardiomyopathy
Genetic mutations and mechanisms in dilated cardiomyopathy Elizabeth M. McNally, … , Jessica R. Golbus, Megan J. Puckelwartz J Clin Invest. 2013;123(1):19-26. https://doi.org/10.1172/JCI62862. Review Series Genetic mutations account for a significant percentage of cardiomyopathies, which are a leading cause of congestive heart failure. In hypertrophic cardiomyopathy (HCM), cardiac output is limited by the thickened myocardium through impaired filling and outflow. Mutations in the genes encoding the thick filament components myosin heavy chain and myosin binding protein C (MYH7 and MYBPC3) together explain 75% of inherited HCMs, leading to the observation that HCM is a disease of the sarcomere. Many mutations are “private” or rare variants, often unique to families. In contrast, dilated cardiomyopathy (DCM) is far more genetically heterogeneous, with mutations in genes encoding cytoskeletal, nucleoskeletal, mitochondrial, and calcium-handling proteins. DCM is characterized by enlarged ventricular dimensions and impaired systolic and diastolic function. Private mutations account for most DCMs, with few hotspots or recurring mutations. More than 50 single genes are linked to inherited DCM, including many genes that also link to HCM. Relatively few clinical clues guide the diagnosis of inherited DCM, but emerging evidence supports the use of genetic testing to identify those patients at risk for faster disease progression, congestive heart failure, and arrhythmia. Find the latest version: https://jci.me/62862/pdf Review series Genetic mutations and mechanisms in dilated cardiomyopathy Elizabeth M. McNally, Jessica R. Golbus, and Megan J. Puckelwartz Department of Human Genetics, University of Chicago, Chicago, Illinois, USA. Genetic mutations account for a significant percentage of cardiomyopathies, which are a leading cause of conges- tive heart failure. -

Epithelial Ovarian Cancer: Searching for New Modulators of Drug Resistance
UNIVERSITÀ DEGLI STUDI DI UDINE Dipartimento di scienze mediche e biologiche PhD COURSE IN BIOMEDICAL SCIENCES AND BIOTECHNOLOGY XXIX cycle PhD THESIS Epithelial Ovarian Cancer: searching for new modulators of drug resistance. PhD student Supervisor Prof. Carlo Ennio Michele Pucillo Valentina Ranzuglia Co-supervisors Dr. Gustavo Baldassarre Dr. Monica Schiappacassi PhD coordinator Prof. Claudio Brancolini Academic year 2015/2016 This PhD work was done at Centro di Riferimento Oncologico (CRO, National Cancer Institute ) of Aviano, in the Division of Experimental Oncology 2 directed by Dr. Gustavo Baldassarre. i Table of contents Abstract 3 1. Introduction 4 1.1 Epithelial Ovarian Cancer 4 1.1.1 Platinum resistance 5 1.2 Functional Genomic Screening 8 1.3 Serum and glucocorticoid-regulated kinases 9 1.3.1 SGK structure 10 1.3.2 SGK activation and regulation 11 1.3.3 Role of SGKs in regulation of molecular and cellular functions 13 1.3.4 Serum- and glucocorticoid-regulated kinases in cancer 15 1.4 Autophagy 17 2.Aim of the study 21 3.Results 22 3.1 Identification of SGK2 as a mediator of platinum sensitivity in EOC cells. 22 3.2 SGK2 silencing sensitizes EOC cells to platinum. 23 3.3 SGK2 overexpression confers an increased resistance to platinum drug. 25 3.4 SGK2-overexpressing cells present an increased in vitro and in vivo growth 26 rate 3.5 SGK2 kinase activity is involved in EOC sensitivity to platinum treatment. 28 3.6 The SGK1/SGK2 kinase inhibitor, GSK650394, reduces cell viability in 29 combination with platinum. 3.7 GSK650394 is able to make SGK2-expressing cells more sensitive to platinum. -

AGC Kinases in Mtor Signaling, in Mike Hall and Fuyuhiko Tamanoi: the Enzymes, Vol
Provided for non-commercial research and educational use only. Not for reproduction, distribution or commercial use. This chapter was originally published in the book, The Enzymes, Vol .27, published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who know you, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial From: ESTELA JACINTO, AGC Kinases in mTOR Signaling, In Mike Hall and Fuyuhiko Tamanoi: The Enzymes, Vol. 27, Burlington: Academic Press, 2010, pp.101-128. ISBN: 978-0-12-381539-2, © Copyright 2010 Elsevier Inc, Academic Press. Author's personal copy 7 AGC Kinases in mTOR Signaling ESTELA JACINTO Department of Physiology and Biophysics UMDNJ-Robert Wood Johnson Medical School, Piscataway New Jersey, USA I. Abstract The mammalian target of rapamycin (mTOR), a protein kinase with homology to lipid kinases, orchestrates cellular responses to growth and stress signals. Various extracellular and intracellular inputs to mTOR are known. mTOR processes these inputs as part of two mTOR protein com- plexes, mTORC1 or mTORC2. Surprisingly, despite the many cellular functions that are linked to mTOR, there are very few direct mTOR substrates identified to date. -

Discovery of the Novel Autophagy Inhibitor Aumitin That Targets Mitochondrial Complex I
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2018 Discovery of the novel autophagy inhibitor Aumitin that targets mitochondrial complex I Lucas Robkea,b,c, Yushi Futamurad, Georgios Konstantinidise, Julian Wilkea,b, Harumi Aonod, Zhwan Mahmoudb, Nobumoto Watanabec,f, Yao-Wen Wue, Hiroyuki Osadac,d, Luca Laraiaa,g *, Herbert Waldmanna,b * a: Max-Planck-Institute of Molecular Physiology, department of Chemical Biology, Otto-Hahn-Str. 11, 44227 Dortmund (Germany); b: Faculty of Chemistry and Chemical Biology, TU Dortmund University, Otto-Hahn-Str. 4a, 44227 Dortmund (Germany); c: RIKEN-Max Planck Joint Research Division for Systems Chemical Biology, RIKEN CSRS, 2-1, Hirosawa, Wako, Saitama 351-0198 (Japan); d: Chemical Biology Research Group, RIKEN CSRS, 2-1, Hirosawa, Wako, Saitama 351-0198 (Japan); e: Chemical Genomics Centre of the Max-Planck-Society, Otto- Hahn-Str. 15, 44227 Dortmund (Germany); f: Bio-Active Compounds Discovery Research Unit, RIKEN CSRS, 2-1, Hirosawa, Wako, Saitama 351-0198 (Japan). g: present address: Department of Chemistry, Technical University of Denmark, Kemitorvet Building 207, Room 124, 2800 Kgs. Lyngby, Denmark. * [email protected], [email protected] SI-Table 1: Structure activity relationship of the di-aminopyrimidines. Starvation = starvation induced autophagy assay; Rapamycin = Rapamycin induced autophagy assay; Viability = survival assessed by means of an ADP-glow assay. > 10 = no inhibition at a test concentration of 10 -
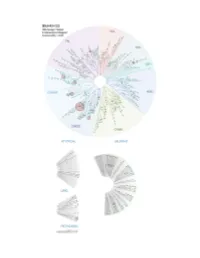
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Kinase Profiling Book
Custom and Pre-Selected Kinase Prof iling to f it your Budget and Needs! As of July 1, 2021 19.8653 mm 128 196 12 Tyrosine Serine/Threonine Lipid Kinases Kinases Kinases Carna Biosciences, Inc. 2007 Carna Biosciences, Inc. Profiling Assays available from Carna Biosciences, Inc. As of July 1, 2021 Page Kinase Name Assay Platform Page Kinase Name Assay Platform 4 ABL(ABL1) MSA 21 EGFR[T790M/C797S/L858R] MSA 4 ABL(ABL1)[E255K] MSA 21 EGFR[T790M/L858R] MSA 4 ABL(ABL1)[T315I] MSA 21 EPHA1 MSA 4 ACK(TNK2) MSA 21 EPHA2 MSA 4 AKT1 MSA 21 EPHA3 MSA 5 AKT2 MSA 22 EPHA4 MSA 5 AKT3 MSA 22 EPHA5 MSA 5 ALK MSA 22 EPHA6 MSA 5 ALK[C1156Y] MSA 22 EPHA7 MSA 5 ALK[F1174L] MSA 22 EPHA8 MSA 6 ALK[G1202R] MSA 23 EPHB1 MSA 6 ALK[G1269A] MSA 23 EPHB2 MSA 6 ALK[L1196M] MSA 23 EPHB3 MSA 6 ALK[R1275Q] MSA 23 EPHB4 MSA 6 ALK[T1151_L1152insT] MSA 23 Erk1(MAPK3) MSA 7 EML4-ALK MSA 24 Erk2(MAPK1) MSA 7 NPM1-ALK MSA 24 Erk5(MAPK7) MSA 7 AMPKα1/β1/γ1(PRKAA1/B1/G1) MSA 24 FAK(PTK2) MSA 7 AMPKα2/β1/γ1(PRKAA2/B1/G1) MSA 24 FER MSA 7 ARG(ABL2) MSA 24 FES MSA 8 AurA(AURKA) MSA 25 FGFR1 MSA 8 AurA(AURKA)/TPX2 MSA 25 FGFR1[V561M] MSA 8 AurB(AURKB)/INCENP MSA 25 FGFR2 MSA 8 AurC(AURKC) MSA 25 FGFR2[V564I] MSA 8 AXL MSA 25 FGFR3 MSA 9 BLK MSA 26 FGFR3[K650E] MSA 9 BMX MSA 26 FGFR3[K650M] MSA 9 BRK(PTK6) MSA 26 FGFR3[V555L] MSA 9 BRSK1 MSA 26 FGFR3[V555M] MSA 9 BRSK2 MSA 26 FGFR4 MSA 10 BTK MSA 27 FGFR4[N535K] MSA 10 BTK[C481S] MSA 27 FGFR4[V550E] MSA 10 BUB1/BUB3 MSA 27 FGFR4[V550L] MSA 10 CaMK1α(CAMK1) MSA 27 FGR MSA 10 CaMK1δ(CAMK1D) MSA 27 FLT1 MSA 11 CaMK2α(CAMK2A) MSA 28 -

Targeting Myddosome Signaling in Waldenström’S
Author Manuscript Published OnlineFirst on August 20, 2018; DOI: 10.1158/1078-0432.CCR-17-3265 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. IRAK 1/4 Inhibition in Waldenström’s Ni, H. et al. Targeting Myddosome Signaling in Waldenström’s Macroglobulinemia with the Interleukin-1 Receptor- associated Kinase 1/4 Inhibitor R191 Haiwen Ni1,2,#, Fazal Shirazi2,#, Veerabhadran Baladandayuthapani3, Heather Lin3, Isere Kuiatse2, Hua Wang2, Richard J. Jones2, Zuzana Berkova2, Yasumichi Hitoshi4, Stephen M. Ansell5, Steven P. Treon6, Sheeba K. Thomas2, Hans C. Lee2, Zhiqiang Wang2, R. Eric Davis2, and Robert Z. Orlowski2,7,* 1Department of Hematology, The Affiliated Hospital of Nanjing University of Traditional Chinese Medicine, Nanjing, JangSu, China; 2Department of Lymphoma and Myeloma, The University of Texas MD Anderson Cancer Center, Houston, TX; 3Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston, TX; 4Rigel, South San Francisco, CA; The 5Division of Hematology, Mayo Clinic, Rochester, MN; The 6Dana Farber Cancer Institute, Harvard Medical School, Boston, MA. 7Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, TX #Indicates that these authors contributed equally. Address correspondence to: Dr. Robert Z. Orlowski, The University of Texas MD Anderson Cancer Center, Department of Lymphoma and Myeloma, 1515 Holcombe Blvd., Unit 429, Houston, TX 77030-4009, E-mail: [email protected], Telephone 713-794-3234, Fax 713- 563-5067 Page 1 Downloaded from clincancerres.aacrjournals.org on September 24, 2021. © 2018 American Association for Cancer Research. Author Manuscript Published OnlineFirst on August 20, 2018; DOI: 10.1158/1078-0432.CCR-17-3265 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -

Kinaseseeker™ Full-Length Panel (112 Wild-Type Kinases)
KinaseSeeker™ Full-Length Panel (112 Wild-Type Kinases) Kinase Group Kinase Group ABL1 full-length TK DDR1 intracellular module TK ACVR1 intracellular module TKL DDR2 intracellular module TK AKT1 full-length AGC EGFR intracellular module TK AKT2 full-length AGC EPHA1 intracellular module TK AKT3 full-length AGC EPHA2 intracellular module TK AMPKa1 full-length CAMK EPHA3 intracellular module TK BLK full-length TK EPHA4 intracellular module TK BTK full-length TK EPHA5 intracellular module TK CAMK1D full-length CAMK EPHA6 intracellular module TK CAMK1G full-length CAMK EPHA7 intracellular module TK CAMK2A full-length CAMK EPHA8 intracellular module TK CAMK2B full-length CAMK EPHB3 intracellular module TK CAMK2D full-length CAMK EPHB4 intracellular module TK CAMK2G full-length CAMK ERBB2 intracellular module TK CAMKK1 full-length Other ERBB4 intracellular module TK CAMKK2 full-length Other FAK full-length TK CASK full-length CAMK FGFR2 intracellular module TK CDKL5 full-length CMGC FGFR3 intracellular module TK CK1d full-length CK1 FGR full-length TK CLK1 full-length CMGC FLT1 intracellular module TK CLK2 full-length CMGC FLT2 intracellular module TK CLK3 full-length CMGC FLT4 intracellular module TK CSF1R intracellular module TK FRK full-length TK CSK full-length TK FYN full-length TK DAPK1 full-length CAMK GRK7 full-length AGC Legend: Full-Length: Construct contains Full-length kinase Intracellular Module: Construct contains Cytoplasmic Region in Receptor Tyrosine Kinases Page 1 of 3 KinaseSeeker™ Full-Length Panel (112 Wild-Type Kinases)