Repositioning the Bioactive Compounds Isolated from Bauhinia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Vascular Plant Survey of Vwaza Marsh Wildlife Reserve, Malawi
YIKA-VWAZA TRUST RESEARCH STUDY REPORT N (2017/18) Vascular Plant Survey of Vwaza Marsh Wildlife Reserve, Malawi By Sopani Sichinga ([email protected]) September , 2019 ABSTRACT In 2018 – 19, a survey on vascular plants was conducted in Vwaza Marsh Wildlife Reserve. The reserve is located in the north-western Malawi, covering an area of about 986 km2. Based on this survey, a total of 461 species from 76 families were recorded (i.e. 454 Angiosperms and 7 Pteridophyta). Of the total species recorded, 19 are exotics (of which 4 are reported to be invasive) while 1 species is considered threatened. The most dominant families were Fabaceae (80 species representing 17. 4%), Poaceae (53 species representing 11.5%), Rubiaceae (27 species representing 5.9 %), and Euphorbiaceae (24 species representing 5.2%). The annotated checklist includes scientific names, habit, habitat types and IUCN Red List status and is presented in section 5. i ACKNOLEDGEMENTS First and foremost, let me thank the Nyika–Vwaza Trust (UK) for funding this work. Without their financial support, this work would have not been materialized. The Department of National Parks and Wildlife (DNPW) Malawi through its Regional Office (N) is also thanked for the logistical support and accommodation throughout the entire study. Special thanks are due to my supervisor - Mr. George Zwide Nxumayo for his invaluable guidance. Mr. Thom McShane should also be thanked in a special way for sharing me some information, and sending me some documents about Vwaza which have contributed a lot to the success of this work. I extend my sincere thanks to the Vwaza Research Unit team for their assistance, especially during the field work. -

Plant Common Name Scientific Name Description of Plant Picture of Plant
Plant common name Description of Plant Picture of Plant Scientific name Strangler Fig The Strangler Fig begins life as a small vine-like plant Ficus thonningii that climbs the nearest large tree and then thickens, produces a branching set of buttressing aerial roots, and strangles its host tree. An easy way to tell the difference between Strangle Figs and other common figs is that the bottom half of the Strangler is gnarled and twisted where it used to be attached to its host, the upper half smooth. A common tree on kopjes and along rivers in Serengeti; two massive Fig trees near Serengeti; the "Tree Where Man was Born" in southern Loliondo, and the "Ancestor Tree" near Endulin, in Ngorongoro are significant for the local Maasai peoples. Wild Date Palm Palms are monocotyledons, the veins in their leaves Phoenix reclinata are parallel and unbranched, and are thus relatives of grasses, lilies, bananas and orchids. The wild Date Palm is the most common of the native palm trees, occurring along rivers and in swamps. The fruits are edible, though horrible tasting, while the thick, sugary sap is made into Palm wine. The tree offers a pleasant, softly rustling, fragrant-smelling shade; the sort of shade you will need to rest in if you try the wine. Candelabra The Candelabra tree is a common tree in the western Euphorbia and Northern parts of Serengeti. Like all Euphorbias, Euphorbia the Candelabra breaks easily and is full of white, candelabrum extremely toxic latex. One drop of this latex can blind or burn the skin. -

Approved Plant List 10/04/12
FLORIDA The best time to plant a tree is 20 years ago, the second best time to plant a tree is today. City of Sunrise Approved Plant List 10/04/12 Appendix A 10/4/12 APPROVED PLANT LIST FOR SINGLE FAMILY HOMES SG xx Slow Growing “xx” = minimum height in Small Mature tree height of less than 20 feet at time of planting feet OH Trees adjacent to overhead power lines Medium Mature tree height of between 21 – 40 feet U Trees within Utility Easements Large Mature tree height greater than 41 N Not acceptable for use as a replacement feet * Native Florida Species Varies Mature tree height depends on variety Mature size information based on Betrock’s Florida Landscape Plants Published 2001 GROUP “A” TREES Common Name Botanical Name Uses Mature Tree Size Avocado Persea Americana L Bahama Strongbark Bourreria orata * U, SG 6 S Bald Cypress Taxodium distichum * L Black Olive Shady Bucida buceras ‘Shady Lady’ L Lady Black Olive Bucida buceras L Brazil Beautyleaf Calophyllum brasiliense L Blolly Guapira discolor* M Bridalveil Tree Caesalpinia granadillo M Bulnesia Bulnesia arboria M Cinnecord Acacia choriophylla * U, SG 6 S Group ‘A’ Plant List for Single Family Homes Common Name Botanical Name Uses Mature Tree Size Citrus: Lemon, Citrus spp. OH S (except orange, Lime ect. Grapefruit) Citrus: Grapefruit Citrus paradisi M Trees Copperpod Peltophorum pterocarpum L Fiddlewood Citharexylum fruticosum * U, SG 8 S Floss Silk Tree Chorisia speciosa L Golden – Shower Cassia fistula L Green Buttonwood Conocarpus erectus * L Gumbo Limbo Bursera simaruba * L -

NABRO Ecological Analysts CC Natural Asset and Botanical Resource Ordinations Environmental Consultants & Wildlife Specialists
NABRO Ecological Analysts CC Natural Asset and Botanical Resource Ordinations Environmental Consultants & Wildlife Specialists ENVIRONMENTAL BASELINE REPORT FOR HANS HOHEISEN WILDLIFE RESEARCH STATION Compiled by Ben Orban, PriSciNat. June 2013 NABRO Ecological Analysts CC. - Reg No: 16549023 / PO Box 11644, Hatfield, Pretoria. Our reference: NABRO / HHWRS/V01 NABRO Ecological Analysts CC Natural Asset and Botanical Resource Ordinations Environmental Consultants & Wildlife Specialists CONTENTS 1 SPECIALIST INVESTIGATORS ............................................................................... 3 2 DECLARATION ............................................................................................................ 3 3 INTRODUCTION ......................................................................................................... 3 4 LOCALITY OF STUDY AREA .................................................................................... 4 4.1 Location ................................................................................................................... 4 5 INFRASTRUCTURE ..................................................................................................... 4 5.1 Fencing ..................................................................................................................... 4 5.2 Camps ...................................................................................................................... 4 5.3 Buildings ................................................................................................................ -

Albuca Spiralis
Flowering Plants of Africa A magazine containing colour plates with descriptions of flowering plants of Africa and neighbouring islands Edited by G. Germishuizen with assistance of E. du Plessis and G.S. Condy Volume 62 Pretoria 2011 Editorial Board A. Nicholas University of KwaZulu-Natal, Durban, RSA D.A. Snijman South African National Biodiversity Institute, Cape Town, RSA Referees and other co-workers on this volume H.J. Beentje, Royal Botanic Gardens, Kew, UK D. Bridson, Royal Botanic Gardens, Kew, UK P. Burgoyne, South African National Biodiversity Institute, Pretoria, RSA J.E. Burrows, Buffelskloof Nature Reserve & Herbarium, Lydenburg, RSA C.L. Craib, Bryanston, RSA G.D. Duncan, South African National Biodiversity Institute, Cape Town, RSA E. Figueiredo, Department of Plant Science, University of Pretoria, Pretoria, RSA H.F. Glen, South African National Biodiversity Institute, Durban, RSA P. Goldblatt, Missouri Botanical Garden, St Louis, Missouri, USA G. Goodman-Cron, School of Animal, Plant and Environmental Sciences, University of the Witwatersrand, Johannesburg, RSA D.J. Goyder, Royal Botanic Gardens, Kew, UK A. Grobler, South African National Biodiversity Institute, Pretoria, RSA R.R. Klopper, South African National Biodiversity Institute, Pretoria, RSA J. Lavranos, Loulé, Portugal S. Liede-Schumann, Department of Plant Systematics, University of Bayreuth, Bayreuth, Germany J.C. Manning, South African National Biodiversity Institute, Cape Town, RSA A. Nicholas, University of KwaZulu-Natal, Durban, RSA R.B. Nordenstam, Swedish Museum of Natural History, Stockholm, Sweden B.D. Schrire, Royal Botanic Gardens, Kew, UK P. Silveira, University of Aveiro, Aveiro, Portugal H. Steyn, South African National Biodiversity Institute, Pretoria, RSA P. Tilney, University of Johannesburg, Johannesburg, RSA E.J. -

Comparison of Seed and Ovule Development in Representative Taxa of the Tribe Cercideae (Caesalpinioideae, Leguminosae) Seanna Reilly Rugenstein Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1983 Comparison of seed and ovule development in representative taxa of the tribe Cercideae (Caesalpinioideae, Leguminosae) Seanna Reilly Rugenstein Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Botany Commons Recommended Citation Rugenstein, Seanna Reilly, "Comparison of seed and ovule development in representative taxa of the tribe Cercideae (Caesalpinioideae, Leguminosae) " (1983). Retrospective Theses and Dissertations. 8435. https://lib.dr.iastate.edu/rtd/8435 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This reproduction was made from a copy of a document sent to us for microfilming. While the most advanced technology has been used to photograph and reproduce this document, the quality of the reproduction is heavily dependent upon the quality of the material submitted. The following explanation of techniques is provided to help clarify markings or notations which may appear on this reproduction. 1. The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting through an image and duplicating adjacent pages to assure complete continuity. 2. -

Choosing Flowering Shrubs for Landscape Color
A WORD OR TWO ABOUT GARDENING Staying Green while Conserving Water in Miami-Dade: Choosing Flowering Shrubs for Landscape Color John McLaughlin1 Introduction Miami-Dade like many other Florida counties is committed to achieving more effective use of available water resources. Reducing household water consumption is a key component of achieving this goal. In Florida it has What is a drought? There is no standard definition but it is more been estimated that from 30-60% of total than rainfall totals. Both place and time have residential water consumption is used to irrigate to be taken into consideration. What defines landscapes. Achieving reductions in landscape drought conditions in Miami differs from those in Las Vegas. Extended periods of water use requires adopting more efficient scant rainfall are expected in Miami during irrigation practices and increasing the proportion of January but not June. A drought is usually drought tolerant plants in area landscapes. recognized when water shortages are sufficient to cause noticeable crop damage, Drought tolerance alone is not sufficient. The trees and/or impact supply. Factors include the and shrubs used in local landscapes also have to immediate period without rain plus the withstand the rainy season (mid-May to mid- actual water deficit (a measure of the October) when Miami-Dade receives most of its’ reduced amount stored in aquifers and surface bodies of water). annual ~60” of rain. For this reason many drought tolerant plants endemic to Mediterranean type climates (warm wet winters and hot dry summers) or year round aridity may not succeed in local landscapes. More likely to adapt to South Florida conditions are flowering shrubs and trees from areas that experience a similar seasonally wet/dry climate (warm dry winters/hot humid summers). -

Mun-Ya-Wana Conservancy
Mun-Ya-Wana Conservancy KwaZulu-Natal South Africa Protected Area Management Plan AUTHORISATION This Management Plan for Mun-Ya-Wana Conservancy is approved: TITLE NAME SIGNATURE AND DATE KwaZulu-Natal MEC: Department of Economic Development, Tourism and Environmental Affairs RECOMMENDED This Management Plan for Mun-Ya-Wana Conservancy is recommended for approval by: TITLE NAME SIGNATURE AND DATE Chief Executive Officer: Ezemvelo KZN Wildlife Chairperson: Biodiversity Conservation Operations Management Committee Management Authority Prepared by 45 Ridge Road Howick P O Box 14310 HOWICK 3290 Tel: 082 804 4412 Email: [email protected] Citation Martindale, G., and Naylor, S. (2018) Mun-Ya-Wana Conservancy Management Plan. Version 1.0. TABLE OF CONTENTS AUTHORISATION TABLE OF CONTENTS LIST OF TABLES LIST OF FIGURES ABBREVIATIONS 1) BACKGROUND 1 1.1 Purpose of the plan 1 1.2 Structure of the plan 3 1.3 Alignment with METT 3 1.4 Introduction 4 1.5 The values of Mun-Ya-Wana Conservancy 5 1.6 Adaptive management 7 2) DESCRIPTION OF MUN-YA-WANA CONSERVANCY AND ITS CONTEXT 8 2.1 The history of Mun-Ya-Wana Conservancy 8 2.2 The legal context for the management of Mun-Ya-Wana Conservancy 12 2.3 Ecological context of Mun-Ya-Wana Conservancy 14 2.4 Cultural and heritage context of Mun-Ya-Wana Conservancy 34 2.5 Socio-economic role of Mun-Ya-Wana Conservancy 35 2.6 The regional and local planning context of Mun-Ya-Wana Conservancy 39 2.7 Operational management within Mun-Ya-Wana Conservancy 43 2.8 Management effectiveness in Mun-Ya-Wana -
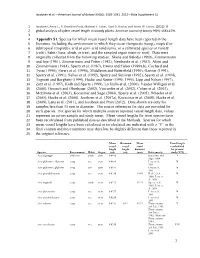
Appendix S1. Species for Which Mean Vessel Length Data Have Been
Jacobsen et al.—American Journal of Botany 99(10): 1583-1591. 2012—Data Supplement S1 1 Jacobsen, Anna L., R. Brandon Pratt, Michael F. Tobin, Uwe G. Hacke, and Frank W. Ewers. (2012). A 2 global analysis of xylem vessel length in woody plants. American Journal of Botany 99(9): 1583-1591. 3 4 Appendix S1. Species for which mean vessel length data have been reported in the 5 literature, including the environment in which they occur (temperate (temp), tropical or 6 subtropical (trop/sub), arid or semi-arid (arid/semi), or a cultivated species or variety 7 (cult)), habit (liana, shrub, or tree), and the sampled organ (stem or root). Data were 8 originally collected from the following studies: Skene and Balodis (1968), Zimmermann 9 and Jeje (1981), Zimmermann and Potter (1982), Newbanks et al. (1983), Aloni and 10 Zimmermann (1984), Sperry et al. (1987), Ewers and Fisher (1989a,b), Cochard and 11 Tyree (1990), Ewers et al. (1990), Middleton and Butterfield (1990), Gartner (1991), 12 Sperry et al. (1991), Salleo et al. (1992), Sperry and Sullivan (1992), Sperry et al. (1994), 13 Tognetti and Borghetti (1994), Hacke and Sauter (1995, 1996), Lipp and Nilsen (1997), 14 Zotz et al. (1997), Kolb and Sperry (1999), Lo Gullo et al. (2000), Vander Willigen et al. 15 (2000), Gorsuch and Oberbauer (2002), Vercambe et al. (2002), Cohen et al. (2003), 16 McElrone et al. (2003), Kocacinar and Sage (2004), Sperry et al. (2005), Wheeler et al. 17 (2005), Hacke et al. (2006), Jacobsen et al. (2007a), Kocacinar et al. (2008), Hacke et al. -

TURF REPLACEMENT PROGRAM MMWD LYL Approved Plant List
LANDSCAPE YOUR LAWN (LYL) TURF REPLACEMENT PROGRAM MMWD LYL Approved Plant List Attached is the current MMWD list of approved plants for the The values are obtained by determining the area of a circle using Landscape Your Lawn (LYL) Program. the plant spread or width as the diameter. To find the area of a circle, square the diameter and multiply by .7854. Squaring the This list is taken from the Water Use Classification of Landscape diameter means multiplying the diameter by itself. For example, a Species (WUCOLS IV) – a widely accepted and commonly used plant with a 5 foot spread would be calculated as follows: source of information on landscape plant water needs. Plants that .7854 x 5 ft diameter x 5 ft diameter = 20 sq ft (values are rounded are listed in WUCOLS IV as “low” or “very low” water use for the Bay to the nearest whole number). Area have been included on this list. However, plants that are considered invasive and are found on the MMWD Invasive Plant List For values not provided, please refer to reputable gardening books are not included in this list and will not be allowed for the LYL or nurseries in order to determine the diameter of the plant at program. maturity, or conduct an internet search using the botanical name and “mature size”. Any plants used in turf conversion that are not on this plant list will not count toward the 50 percent plant coverage requirement nor CA Natives will they be eligible for a rebate under LYL Option 1. Native plants are perfectly suited to our climate, soil, and animals. -

WUCOLS List S Abelia Chinensis Chinese Abelia M ? ? M / / Copyright © UC Regents, Davis Campus
Ba Bu G Gc P Pm S Su T V N Botanical Name Common Name 1 2 3 4 5 6 Symbol Vegetation Used in Type WUCOLS List S Abelia chinensis Chinese abelia M ? ? M / / Copyright © UC Regents, Davis campus. All rights reserved. bamboo Ba S Abelia floribunda Mexican abelia M ? M M / / S Abelia mosanensis 'Fragrant Abelia' fragrant abelia ? ? ? ? ? ? bulb Bu S Abelia parvifolia (A. longituba) Schuman abelia ? ? ? M ? ? grass G groundcover GC Gc S Abelia x grandiflora and cvs. glossy abelia M M M M M / perennial* P S Abeliophyllum distichum forsythia M M ? ? ? ? palm and cycad Pm S Abelmoschus manihot (Hibiscus manihot) sunset muskmallow ? ? ? L ? ? T Abies pinsapo Spanish fir L L L / / / shrub S succulent Su T N Abies spp. (CA native and non-native) fir M M M M / / P N Abronia latifolia yellow sand verbena VL VL VL / ? ? tree T P N Abronia maritima sand verbena VL VL VL / ? ? vine V California N native S N Abutilon palmeri Indian mallow L L L L M M S Abutilon pictum thompsonii variegated Chinese lantern M H M M ? ? Sunset WUCOLS CIMIS ET Representative Number climate 0 Region zones** Cities zones* S Abutilon vitifolium flowering maple M M M / ? ? Healdsburg, Napa, North- San Jose, Salinas, Central 14, 15, 16, 17 1, 2, 3, 4, 6, 8 San Francisco, Coastal San Luis Obispo S Abutilon x hybridum & cvs. flowering maple M H M M / / 1 Auburn, Central Bakersfield, Chico, 8, 9, 14 12, 14, 15, 16 Valley Fresno, Modesto, Sacramento S T Acacia abyssinica Abyssinian acacia / ? / ? / L 2 Irvine, Los South Angeles, Santa 22, 23, 24 1, 2, 4, 6 Coastal Barbara, Ventura, -

The Whole and the Parts: Relationships Between Floral Architecture and Floral Organ Shape, and Their Repercussions on the Interpretation of Fragmentary Floral Fossils
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2008 The whole and the parts: relationships between floral architecture and floral organ shape, and their repercussions on the interpretation of fragmentary floral fossils Endress, P K Abstract: Floral architecture and floral organ shape are interrelated to some extent as can be seen inthe diversity of extant angiosperm groups. The shape of fragmentary fossil material, such as single organs, may therefore give hints for the reconstruction of the architecture of a flower. This study is partly a review and partly provides original material and new points of view on organ-architecture interrelationships. Several topics are illustrated with examples: (1) autonomous and imprinted shape, exemplified by cuneate organs, especially stamens; (2) conditions for valvate anther dehiscence; (3) lability in number and shape of reduced organs that have decreased in size and lost their original function; (4) long hairs as filling material of irregular spaces; (5) architectural conditions for the presence of orthotropous ovules; (6) structural differences between exposed and covered organ parts in bud; and (7) sepal aestivation and petal elaboration. DOI: https://doi.org/10.3417/2006190 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-11688 Journal Article Published Version Originally published at: Endress, P K (2008). The whole and the parts: relationships between floral architecture and floral organ shape, and their repercussions on the interpretation of fragmentary floral fossils. Annals of the Missouri Botanical Garden, 95(1):101-120.