Isolation and Characterization of Microsatellite Loci in Castanopsis Fissa in Lower Subtropical China
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lepidoptera, Incurvariidae) with Two New Species from China and Japan
Zootaxa 4927 (2): 209–233 ISSN 1175-5326 (print edition) https://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2021 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4927.2.3 http://zoobank.org/urn:lsid:zoobank.org:pub:96B9981B-01B5-4828-A4C6-E2E4A08DB8F2 Review of the genus Vespina (Lepidoptera, Incurvariidae) with two new species from China and Japan TOSHIYA HIROWATARI1*, SADAHISA YAGI1, ISSEI OHSHIMA2, GUO-HUA HUANG3 & MIN WANG4 1Entomological laboratory, Faculty of Agriculture, Kyushu University, Fukuoka, 819-0395 Japan. [email protected]; https://orcid.org/0000-0002-4261-1219 2Department of Life and Environmental Sciences, Kyoto Prefectural University, Kyoto, 606-8522 Japan. [email protected]; https://orcid.org/0000-0001-8295-9749 3Hunan Provincial Key Laboratory for Biology and Control of Plant Diseases and Insect Pests, Hunan Agricultural University, Changsha 410128, Hunan, China. [email protected]; https://orcid.org/0000-0002-6841-0095 4Department of Entomology, South China Agricultural University, Guangzhou 510640, Guangdong, China. [email protected]; https://orcid.org/0000-0001-5834-4058 *Corresponding author. [email protected]; https://orcid.org/0000-0002-6839-2229 Abstract Asian species of the genus Vespina Davis, 1972 (Lepidoptera, Incurvariidae) are mainly reviewed. Vespina meridiana Hirowatari & Yagi sp. nov. from the Ryukyu Islands, Japan, and Vespina sichuana Hirowatari, Huang & Wang sp. nov. from Sichuan, China, are described. The previously known Vespina species are associated with plants from the Fagaceae family on the western coast of the USA and East Asia and with Sapindaceae (Aceraceae) in eastern Europe. -

Study on the Genetic Structure Based on Geographic Populations of the Endangered Tree Species: Liriodendron Chinense
Article Study on the Genetic Structure Based on Geographic Populations of the Endangered Tree Species: Liriodendron chinense Peng-Yan Zhou 1,† , Li-Xing Hui 1,†, Shu-Jing Huang 1, Zhou-Xian Ni 1 , Fa-Xin Yu 2 and Li-An Xu 1,* 1 Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing 210037, China; [email protected] (P.-Y.Z.); [email protected] (L.-X.H.); [email protected] (S.-J.H.); [email protected] (Z.-X.N.) 2 The Key Laboratory of Horticultural Plant Genetic and Improvement of Jiangxi Province, Institute of Biological Resources, Jiangxi Academy of Sciences, Nanchang 330096, China; [email protected] * Correspondence: [email protected]; Tel.: +86-025-8542-7882 † Peng-Yan Zhou and Li-Xing Hui contributed equal. Abstract: Liriodendron chinense (Hemsley) Sargent is a Class II protected plant in China as natural populations are on the verge of extinction. There is still a lack of systematic research on the genetic resources of its geographic populations. In this study, we used 20 pairs of SSR markers with high polymorphism to analyze a total of 808 L. chinense samples from 22 regions, and 63 Liriodendron tulipifera Linn samples from 2 regions were used as a comparison group. The results revealed a total of 78 alleles in L. chinense, and the average expected heterozygosity (He) was 0.558, showing a low level of genetic diversity. The degree of differentiation of L. chinense was high, with the differentiation coefficient (Fst) as high as 0.302, which is related to the low gene flow (Nm = 0.578). -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m. -

Report of Rapid Biodiversity Assessments at Cenwanglaoshan Nature Reserve, Northwest Guangxi, China, 1999 and 2002
Report of Rapid Biodiversity Assessments at Cenwanglaoshan Nature Reserve, Northwest Guangxi, China, 1999 and 2002 Kadoorie Farm and Botanic Garden in collaboration with Guangxi Zhuang Autonomous Region Forestry Department Guangxi Forestry Survey and Planning Institute South China Institute of Botany South China Normal University Institute of Zoology, CAS March 2003 South China Forest Biodiversity Survey Report Series: No. 27 (Online Simplified Version) Report of Rapid Biodiversity Assessments at Cenwanglaoshan Nature Reserve, Northwest Guangxi, China, 1999 and 2002 Editors John R. Fellowes, Bosco P.L. Chan, Michael W.N. Lau, Ng Sai-Chit and Gloria L.P. Siu Contributors Kadoorie Farm and Botanic Garden: Gloria L.P. Siu (GS) Bosco P.L. Chan (BC) John R. Fellowes (JRF) Michael W.N. Lau (ML) Lee Kwok Shing (LKS) Ng Sai-Chit (NSC) Graham T. Reels (GTR) Roger C. Kendrick (RCK) Guangxi Zhuang Autonomous Region Forestry Department: Xu Zhihong (XZH) Pun Fulin (PFL) Xiao Ma (XM) Zhu Jindao (ZJD) Guangxi Forestry Survey and Planning Institute (Comprehensive Tan Wei Fu (TWF) Planning Branch): Huang Ziping (HZP) Guangxi Natural History Museum: Mo Yunming (MYM) Zhou Tianfu (ZTF) South China Institute of Botany: Chen Binghui (CBH) Huang Xiangxu (HXX) Wang Ruijiang (WRJ) South China Normal University: Li Zhenchang (LZC) Chen Xianglin (CXL) Institute of Zoology CAS (Beijing): Zhang Guoqing (ZGQ) Chen Deniu (CDN) Nanjing University: Chen Jianshou (CJS) Wang Songjie (WSJ) Xinyang Teachers’ College: Li Hongjing (LHJ) Voluntary specialist: Keith D.P. Wilson (KW) Background The present report details the findings of visits to Northwest Guangxi by members of Kadoorie Farm and Botanic Garden (KFBG) in Hong Kong and their colleagues, as part of KFBG's South China Biodiversity Conservation Programme. -

Kadoorie Farm and Botanic Garden, 2004. Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001
Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001 Kadoorie Farm and Botanic Garden in collaboration with Zhongshan University Zhaoqing Forestry Bureau February 2004 South China Forest Biodiversity Survey Report Series: No. 37 (Online Simplified Version) Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001 Editors Bosco P.L. Chan, Ng Sai-Chit, Michael W.N. Lau and John R. Fellowes Contributors Kadoorie Farm and Botanic Garden: Michael W.N. Lau (ML) Bosco P.L. Chan (BC) John R. Fellowes (JRF) Lee Kwok Shing (LKS) Ng Sai-Chit (NSC) Roger Kendrick (RCK) Zhongshan University: Chang Hong (CH) Voluntary specialists: Graham T. Reels (GTR) Keith D.P. Wilson (KW) Background The present report details the findings of a trip to Northwest Guangdong by members of Kadoorie Farm and Botanic Garden (KFBG) in Hong Kong and their colleagues, as part of KFBG's South China Biodiversity Conservation Programme (renamed the China Programme in 2003). The overall aim of the programme is to minimise the loss of forest biodiversity in the region, and the emphasis in the first three years is on gathering up-to-date information on the distribution and status of fauna and flora. Citation Kadoorie Farm and Botanic Garden, 2004. Report of Rapid Biodiversity Assessments at Dachouding and Sanyue Nature Reserves, Northwest Guangdong, China, April 2001 . South China Forest Biodiversity Survey Report Series (Online Simplified Version): No. 37. KFBG, Hong Kong SAR, ii + 33 pp. Copyright Kadoorie Farm and Botanic Garden Corporation Lam Kam Road, Tai Po, N.T., Hong Kong February 2004 - i - Contents Objectives ……………………………………………………………………………………. -

FAGACEAE 1. FAGUS Linnaeus, Sp. Pl. 2: 997. 1753
Flora of China 4: 314–400. 1999. 1 FAGACEAE 壳斗科 qiao dou ke Huang Chengjiu (黄成就 Huang Ching-chieu)1, Zhang Yongtian (张永田 Chang Yong-tian)2; Bruce Bartholomew3 Trees or rarely shrubs, monoecions, evergreen or deciduous. Stipules usually early deciduous. Leaves alternate, sometimes false-whorled in Cyclobalanopsis. Inflorescences unisexual or androgynous with female cupules at the base of an otherwise male inflorescence. Male inflorescences a pendulous head or erect or pendulous catkin, sometimes branched; flowers in dense cymules. Male flower: sepals 4–6(–9), scalelike, connate or distinct; petals absent; filaments filiform; anthers dorsifixed or versatile, opening by longitudinal slits; with or without a rudimentary pistil. Female inflorescences of 1–7 or more flowers subtended individually or collectively by a cupule formed from numerous fused bracts, arranged individually or in small groups along an axis or at base of an androgynous inflorescence or on a separate axis. Female flower: perianth 1–7 or more; pistil 1; ovary inferior, 3–6(– 9)-loculed; style and carpels as many as locules; placentation axile; ovules 2 per locule. Fruit a nut. Seed usually solitary by abortion (but may be more than 1 in Castanea, Castanopsis, Fagus, and Formanodendron), without endosperm; embryo large. Seven to 12 genera (depending on interpretation) and 900–1000 species: worldwide except for tropical and S Africa; seven genera and 294 species (163 endemic, at least three introduced) in China. Many species are important timber trees. Nuts of Fagus, Castanea, and of most Castanopsis species are edible, and oil is extracted from nuts of Fagus. Nuts of most species of this family contain copious amounts of water soluble tannin. -

Identification of a Natural Hybrid Between Castanopsis Sclerophylla
Article Identification of a Natural Hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) Based on Chloroplast and Nuclear DNA Sequences Xiaorong Zeng, Risheng Chen, Yunxin Bian, Xinsheng Qin, Zhuoxin Zhang and Ye Sun * Guangdong Key Laboratory for Innovative Development and Utilization of Forest Plant Germplasm, College of Forestry and Landscape Architecture, South China Agriculture University, Guangzhou 510642, China; [email protected] (X.Z.); [email protected] (R.C.); [email protected] (Y.B.); [email protected] (X.Q.); [email protected] (Z.Z.) * Correspondence: [email protected]; Tel.: +86-136-4265-9676 Received: 30 June 2020; Accepted: 7 August 2020; Published: 11 August 2020 Abstract: Castanopsis kuchugouzhuiHuang et Y. T. Chang was recorded in Flora Reipublicae Popularis × Sinicae (FRPS) as a hybrid species on Yuelushan mountain, but it is treated as a hybrid between Castanopsis sclerophylla (Lindl.) Schott. and Castanopsis tibetana Hance in Flora of China. After a thorough investigation on Yuelushan mountain, we found a population of C. sclerophylla and one individual of C. kuchugouzhui, × but no living individual of C. tibetana. We collected C. kuchugouzhui, and we sampled 42 individuals of × C. sclerophylla from Yuelushan and Xiushui and 43 individuals of C. tibetana from Liangyeshan and Xiushui. Weused chloroplast DNA sequences and 29 nuclear microsatellite markers to investigate if C. kuchugouzhui × is a natural hybrid between C. sclerophylla and C. tibetana. The chloroplast haplotype analysis showed that C. kuchugouzhuishared haplotype H2 with C. sclerophylla on Yuelushan. The STRUCTURE analysis × identified two distinct genetic pools that corresponded well to C. sclerophylla and C. tibetana, revealing the genetic admixture of C. -

Relationships Between Vegetation and Stomata, and Between Vegetation and Pollen Surface Soil in Yunnan, Southwest China
Article Ecology May 2013 Vol.58 No.15: 17751786 doi: 10.1007/s11434-012-5657-2 Relationships between vegetation and stomata, and between vegetation and pollen surface soil in Yunnan, Southwest China SHEN HuaDong1,2, LI ChunHai1*, WAN HeWen3, TONG GuoBang4, LIU JinSong5,6 & DAN Johnson7 1 State Key Laboratory of Lake Science and Environment, Nanjng Institute of Geography and Limnology, Chinese Academy of Sciences, Nanjing 210008, China; 2 University of Chinese Academy of Sciences, Beijing 100049, China; 3 College of Life Sciences, Anhui University, Hefei 230039, China; 4 Institute of Hydrogeology and Environmental Geology, Chinese Academy of Geological Sciences, Zhengding 050803, China; 5 Mathematics and Information Science College, Hebei Normal University, Shijiazhuang 050024, China; 6 The Key Laboratory of Calculation and Application of Hebei Province, Shijiazhuang 050024, China; 7 Environmental Science, Department of Geography, University of Lethbridge, Alberta T1K 3M4, Canada Received August 26, 2012; accepted November 30, 2012; published online February 21, 2013 Surface pollen and stomata of 61 samples collected in a study area ranging from tropical seasonal rainforest to oak forest (Quer- cus spinosa) in the Yulong Snow Mountain region in Yunnan, China, are used to distinguish vegetation communities. The results show that tropical seasonal rainforest (and mountain rainforest), south subtropical evergreen broad-leaved forest, and Quercus shrub are distinguished effectively from other vegetation types by analysis of surface pollen. The south subtropical evergreen broad-leaved forest, Pinus kesiya forest and evergreen broadleaf forest are distinguished effectively from other types of vegetation by pollen analysis. However, P. yunnanensis forest is not distinguished from other vegetation types, and P. -

Supplementary Material
Xiang et al., Page S1 Supporting Information Fig. S1. Examples of the diversity of diaspore shapes in Fagales. Fig. S2. Cladogram of Fagales obtained from the 5-marker data set. Fig. S3. Chronogram of Fagales obtained from analysis of the 5-marker data set in BEAST. Fig. S4. Time scale of major fagalean divergence events during the past 105 Ma. Fig. S5. Confidence intervals of expected clade diversity (log scale) according to age of stem group. Fig. S6. Evolution of diaspores types in Fagales with BiSSE model. Fig. S7. Evolution of diaspores types in Fagales with Mk1 model. Fig. S8. Evolution of dispersal modes in Fagales with MuSSE model. Fig. S9. Evolution of dispersal modes in Fagales with Mk1 model. Fig. S10. Reconstruction of pollination syndromes in Fagales with BiSSE model. Fig. S11. Reconstruction of pollination syndromes in Fagales with Mk1 model. Fig. S12. Reconstruction of habitat shifts in Fagales with MuSSE model. Fig. S13. Reconstruction of habitat shifts in Fagales with Mk1 model. Fig. S14. Stratigraphy of fossil fagalean genera. Table S1 Genera of Fagales indicating the number of recognized and sampled species, nut sizes, habits, pollination modes, and geographic distributions. Table S2 List of taxa included in this study, sources of plant material, and GenBank accession numbers. Table S3 Primers used for amplification and sequencing in this study. Table S4 Fossil age constraints utilized in this study of Fagales diversification. Table S5 Fossil fruits reviewed in this study. Xiang et al., Page S2 Table S6 Statistics from the analyses of the various data sets. Table S7 Estimated ages for all families and genera of Fagales using BEAST. -
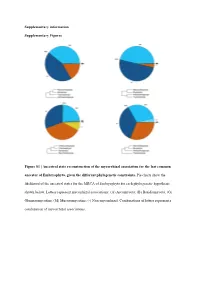
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Species Richness, Forest Types and Regeneration of Schima in the Subtropical Forest Ecosystem of Yunnan, Southwestern China Cindy Q
Tang et al. Forest Ecosystems (2020) 7:35 https://doi.org/10.1186/s40663-020-00244-1 RESEARCH Open Access Species richness, forest types and regeneration of Schima in the subtropical forest ecosystem of Yunnan, southwestern China Cindy Q. Tang1* , Peng-Bin Han1*, Shuaifeng Li2, Li-Qin Shen1, Diao-Shun Huang1, Yun-Fang Li3, Ming-Chun Peng1, Chong-Yun Wang1, Xiao-Shuang Li4, Wei Li1, Wei Wang5 and Zhi-Ying Zhang1 Abstract Background: Schima genus of Theaceae is confined to subtropics and tropics of South, East and Southeast Asia. Thirteen species of Schima are distributed in subtropical China. Many of them appear as dominant canopy species in the subtropical forests. To date, Schima species richness distribution patterns of China have remained unknown. Meanwhile, there has been a longtime debate as to whether forests dominated by Schima species are early or late successional forests. We aim to clarify Schima species richness patterns and these species’ roles in the forest succession and regeneration dynamics of the subtropical ecosystem in Yunnan Province, China. Method: We mapped Schima species richness distribution patterns in China. Based on 71 vegetation plots, we analyzed forest characteristics, population structure, and regeneration dynamics of Schima species in Yunnan. Results: Yunnan was found to harbor the greatest richness and the highest rarity-weighted richness of Schima species in the subtropical regions of China. We classified five primary and six secondary forest types containing Schima species as one of dominants. Yunnan had the high floristic diversity and varying stand structure of forests containing Schima species. The Schima species studied generally had a sporadic regeneration type and a long life- span. -

Taimeselts Fagales Süstemaatika Ja Levik Maailmas
Tartu Ülikool Loodus- ja tehnoloogiateaduskond Ökoloogia ja Maateaduste Instituut Botaanika osakond Hanna Hirve TAIMESELTS FAGALES SÜSTEMAATIKA JA LEVIK MAAILMAS Bakalaureusetöö Juhendaja: professor Urmas Kõljalg Tartu 2014 Sisukord Sisukord ............................................................................................................................ 2 Sissejuhatus ...................................................................................................................... 4 1. Taimeseltsist Fagales üldiselt ................................................................................... 5 2. Takson Betulaceae ................................................................................................... 7 2.1 Iseloomustus ja levik ......................................................................................... 7 2.2 Morfoloogilised tunnused .................................................................................. 8 2.3 Fülogenees ......................................................................................................... 9 2.4 Tähtsus ............................................................................................................... 9 3. Takson Casuarinaceae ............................................................................................ 10 3.1 Iseloomustus ja levik ....................................................................................... 10 3.2 Morfoloogilised tunnused ...............................................................................