Identification of a Natural Hybrid Between Castanopsis Sclerophylla
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Hybrid Vigor Between Native and Introduced Salamanders Raises New Challenges for Conservation
Hybrid vigor between native and introduced salamanders raises new challenges for conservation Benjamin M. Fitzpatrick*† and H. Bradley Shaffer‡ *Ecology and Evolutionary Biology, University of Tennessee, Knoxville, TN 37996; and ‡Evolution and Ecology, University of California, Davis, CA 95616 Edited by John C. Avise, University of California, Irvine, CA, and approved August 10, 2007 (received for review May 22, 2007) Hybridization between differentiated lineages can have many populations by alleviating inbreeding depression (13, 14) or different consequences depending on fitness variation among facilitating adaptive evolution in modified or degraded habitats hybrid offspring. When introduced organisms hybridize with na- (15–17). tives, the ensuing evolutionary dynamics may substantially com- The long-term consequences of hybridization are strongly plicate conservation decisions. Understanding the fitness conse- influenced by the genetic basis of hybrid fitness. In the case of quences of hybridization is an important first step in predicting its hybrid vigor, genetic models fall into two classes: heterozygote evolutionary outcome and conservation impact. Here, we mea- advantage and recombinant hybrid vigor (18–20). Heterozygote sured natural selection caused by differential viability of hybrid advantage (overdominance) refers to beneficial interactions larvae in wild populations where native California Tiger between heterospecific alleles of a single locus. Recombinant Salamanders (Ambystoma californiense) and introduced Barred hybrid vigor depends on multilocus genotypes and may be caused Tiger Salamanders (Ambystoma tigrinum mavortium) have been by epistasis (beneficial interactions between heterospecific al- hybridizing for 50–60 years. We found strong evidence of hybrid leles from different loci) or by complementary effects of inde- vigor; mixed-ancestry genotypes had higher survival rates than pendent advantageous alleles from each parental population (19, genotypes containing mostly native or mostly introduced alleles. -

Quercus Robur (Fagaceae)
Quercus robur (Fagaceae) The map description EEBIO area The integrated map shows the distribution and changes in the areal’s boundaries of pedunculate oak (Quercus robur). Q. robur is the dominant forest- formative species in the belt of broadleaf and mixed 4 needleleaf-broadleaf forests in the plains of the European part of the former USSR (Sokolov et al. 1977). In the northern part of its areal Q. robur grows in river valleys. In the central part, it forms mixed forests with Picea abies; closer to the south – a belt of broadleaf forests where Q. robur dominates. At the areal’s south boundary it forms small (marginal) forests in ravines and flood-plains (Atlas of Areals and Resources… , 1976). Q. robur belongs to the thermophilic species. The low temperature bound of possible occurrence of oak forests is marked by an average annual of 2?C l (http://www.forest.ru – in Russian). Therefore, l l l l l l hypotretically, oak areal boundaries will shift along l ll l l l l l l l with the changes in the average annual temperature. l l l l l l l l l l l For Yearly map of averaged mean annual air l l l l l l l l l l l l l l l l l l lll temperature (Afonin A., Lipiyaynen K., Tsepelev V., l l l l l l l ll ll l l l 2005) see http://www.agroatlas.spb.ru Climate. l l l l l l l l l l l l l l l l Oak forests are of great importance for the water l l l l l ll l lll l l l l l l l l l l l l l l l l l l l regime and soil structure, especially on the steep l l l l l l l l l l l l l l l l l l l l l l l l l l l l l slopes of river valleys and in forest-poor areas. -

5 Fagaceae Trees
CHAPTER 5 5 Fagaceae Trees Antoine Kremerl, Manuela Casasoli2,Teresa ~arreneche~,Catherine Bod6n2s1, Paul Sisco4,Thomas ~ubisiak~,Marta Scalfi6, Stefano Leonardi6,Erica ~akker~,Joukje ~uiteveld', Jeanne ~omero-Seversong, Kathiravetpillai Arumuganathanlo, Jeremy ~eror~',Caroline scotti-~aintagne", Guy Roussell, Maria Evangelista Bertocchil, Christian kxerl2,Ilga porth13, Fred ~ebard'~,Catherine clark15, John carlson16, Christophe Plomionl, Hans-Peter Koelewijn8, and Fiorella villani17 UMR Biodiversiti Genes & Communautis, INRA, 69 Route d'Arcachon, 33612 Cestas, France, e-mail: [email protected] Dipartimento di Biologia Vegetale, Universita "La Sapienza", Piazza A. Moro 5,00185 Rome, Italy Unite de Recherche sur les Especes Fruitikres et la Vigne, INRA, 71 Avenue Edouard Bourlaux, 33883 Villenave d'Ornon, France The American Chestnut Foundation, One Oak Plaza, Suite 308 Asheville, NC 28801, USA Southern Institute of Forest Genetics, USDA-Forest Service, 23332 Highway 67, Saucier, MS 39574-9344, USA Dipartimento di Scienze Ambientali, Universitk di Parma, Parco Area delle Scienze 1lIA, 43100 Parma, Italy Department of Ecology and Evolution, University of Chicago, 5801 South Ellis Avenue, Chicago, IL 60637, USA Alterra Wageningen UR, Centre for Ecosystem Studies, P.O. Box 47,6700 AA Wageningen, The Netherlands Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA lo Flow Cytometry and Imaging Core Laboratory, Benaroya Research Institute at Virginia Mason, 1201 Ninth Avenue, Seattle, WA 98101, -

Transformations of Lamarckism Vienna Series in Theoretical Biology Gerd B
Transformations of Lamarckism Vienna Series in Theoretical Biology Gerd B. M ü ller, G ü nter P. Wagner, and Werner Callebaut, editors The Evolution of Cognition , edited by Cecilia Heyes and Ludwig Huber, 2000 Origination of Organismal Form: Beyond the Gene in Development and Evolutionary Biology , edited by Gerd B. M ü ller and Stuart A. Newman, 2003 Environment, Development, and Evolution: Toward a Synthesis , edited by Brian K. Hall, Roy D. Pearson, and Gerd B. M ü ller, 2004 Evolution of Communication Systems: A Comparative Approach , edited by D. Kimbrough Oller and Ulrike Griebel, 2004 Modularity: Understanding the Development and Evolution of Natural Complex Systems , edited by Werner Callebaut and Diego Rasskin-Gutman, 2005 Compositional Evolution: The Impact of Sex, Symbiosis, and Modularity on the Gradualist Framework of Evolution , by Richard A. Watson, 2006 Biological Emergences: Evolution by Natural Experiment , by Robert G. B. Reid, 2007 Modeling Biology: Structure, Behaviors, Evolution , edited by Manfred D. Laubichler and Gerd B. M ü ller, 2007 Evolution of Communicative Flexibility: Complexity, Creativity, and Adaptability in Human and Animal Communication , edited by Kimbrough D. Oller and Ulrike Griebel, 2008 Functions in Biological and Artifi cial Worlds: Comparative Philosophical Perspectives , edited by Ulrich Krohs and Peter Kroes, 2009 Cognitive Biology: Evolutionary and Developmental Perspectives on Mind, Brain, and Behavior , edited by Luca Tommasi, Mary A. Peterson, and Lynn Nadel, 2009 Innovation in Cultural Systems: Contributions from Evolutionary Anthropology , edited by Michael J. O ’ Brien and Stephen J. Shennan, 2010 The Major Transitions in Evolution Revisited , edited by Brett Calcott and Kim Sterelny, 2011 Transformations of Lamarckism: From Subtle Fluids to Molecular Biology , edited by Snait B. -

Nothofagus, Key Genus of Plant Geography, in Time
Nothofagus, key genus of plant geography, in time and space, living and fossil, ecology and phylogeny C.G.G.J. van Steenis Rijksherbarium, Leyden, Holland Contents Summary 65 1. Introduction 66 New 2. Caledonian species 67 of and Caledonia 3. Altitudinal range Nothofagus in New Guinea New 67 Notes of 4. on distribution Nothofagus species in New Guinea 70 5. Dominance of Nothofagus 71 6. Symbionts of Nothofagus 72 7. Regeneration and germination of Nothofagus in New Guinea 73 8. Dispersal in Nothofagus and its implications for the genesis of its distribution 74 9. The South Pacific and subantarctic climate, present and past 76 10. The fossil record 78 of in time and 11. Phylogeny Nothofagus space 83 12. Bi-hemispheric ranges homologous with that of Fagoideae 89 13. Concluding theses 93 Acknowledgements 95 Bibliography 95 Postscript 97 Summary Data are given on the taxonomy and ecology of the genus. Some New Caledonian in descend the lowland. Details the distri- species grow or to are provided on bution within New Guinea. For dominance of Nothofagus, and Fagaceae in general, it is suggested that this. Some in New possibly symbionts may contribute to notes are made onregeneration and germination Guinea. A is devoted a discussion of which to be with the special chapter to dispersal appears extremely slow, implication that Nothofagus indubitably needs land for its spread, and has needed such for attaining its colossal range, encircling onwards of New Guinea the South Pacific (fossil pollen in Antarctica) to as far as southern South America. Map 1. An is other chapter devoted to response ofNothofagus to the present climate. -

Patterns of Occurrence of Hybrids of Castanopsis Cuspidata and C
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Kanazawa University Repository for Academic Resources Patterns of occurrence of hybrids of Castanopsis cuspidata and C. sieboldii in the IBP Minamata Special Research Area , Kumamoto Prefecture , Japan 著者 Kobayashi Satoshi, Hiroki Shozo journal or 植物地理・分類研究 = The journal of publication title phytogeography and toxonomy volume 51 number 1 page range 63-67 year 2003-06-25 URL http://hdl.handle.net/2297/48538 Journal of Phytogeography and Taxonomy 51 : 63-67, 2003 !The Society for the Study of Phytogeography and Taxonomy 2003 Satoshi Kobayashi and Shozo Hiroki : Patterns of occurrence of hybrids of Castanopsis cuspidata and C. sieboldii in the IBP Minamata Special Research Area , Kumamoto Prefecture , Japan Graduate School of Human Informatics, Nagoya University, Chikusa-Ku, Nagoya 464―8601, Japan Castanopsis cuspidata(Thunb.)Schottky and However, it is difficult to identify the hybrids by C. sieboldii(Makino)Hatus. ex T. Yamaz. et nut morphology alone, because the nut shapes of Mashiba are dominant components of the ever- the 2 species are variable and can overlap with green broad-leaved forests of southwestern Ja- each other. Kobayashi et al.(1998)showed that pan, including parts of Honshu, Kyushu and hybrids have a chimeric structure of both 1 and Shikoku but excluding the Ryukyu Islands(Hat- 2 epidermal layers within a leaf. These morpho- tori and Nakanishi 1983).Although these 2 Cas- logical differences among C. cuspidata, C. sie- tanopsis species are both climax species, it is boldii and their hybrid can be confirmed by ge- very difficult to distinguish them because of the netic differences in nuclear species-specific existence of an intermediate type(hybrid). -

Diversity of Wisconsin Rosids
Diversity of Wisconsin Rosids . oaks, birches, evening primroses . a major group of the woody plants (trees/shrubs) present at your sites The Wind Pollinated Trees • Alternate leaved tree families • Wind pollinated with ament/catkin inflorescences • Nut fruits = 1 seeded, unilocular, indehiscent (example - acorn) *Juglandaceae - walnut family Well known family containing walnuts, hickories, and pecans Only 7 genera and ca. 50 species worldwide, with only 2 genera and 4 species in Wisconsin Carya ovata Juglans cinera shagbark hickory Butternut, white walnut *Juglandaceae - walnut family Leaves pinnately compound, alternate (walnuts have smallest leaflets at tip) Leaves often aromatic from resinous peltate glands; allelopathic to other plants Carya ovata Juglans cinera shagbark hickory Butternut, white walnut *Juglandaceae - walnut family The chambered pith in center of young stems in Juglans (walnuts) separates it from un- chambered pith in Carya (hickories) Juglans regia English walnut *Juglandaceae - walnut family Trees are monoecious Wind pollinated Female flower Male inflorescence Juglans nigra Black walnut *Juglandaceae - walnut family Male flowers apetalous and arranged in pendulous (drooping) catkins or aments on last year’s woody growth Calyx small; each flower with a bract CA 3-6 CO 0 A 3-∞ G 0 Juglans cinera Butternut, white walnut *Juglandaceae - walnut family Female flowers apetalous and terminal Calyx cup-shaped and persistant; 2 stigma feathery; bracted CA (4) CO 0 A 0 G (2-3) Juglans cinera Juglans nigra Butternut, white -

Hybrid Fitness, Adaptation and Evolutionary Diversification: Lessons
Heredity (2012) 108, 159–166 & 2012 Macmillan Publishers Limited All rights reserved 0018-067X/12 www.nature.com/hdy REVIEW Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana Irises ML Arnold, ES Ballerini and AN Brothers Estimates of hybrid fitness have been used as either a platform for testing the potential role of natural hybridization in the evolution of species and species complexes or, alternatively, as a rationale for dismissing hybridization events as being of any evolutionary significance. From the time of Darwin’s publication of The Origin, through the neo-Darwinian synthesis, to the present day, the observation of variability in hybrid fitness has remained a challenge for some models of speciation. Yet, Darwin and others have reported the elevated fitness of hybrid genotypes under certain environmental conditions. In modern scientific terminology, this observation reflects the fact that hybrid genotypes can demonstrate genotypeÂenvironment interactions. In the current review, we illustrate the development of one plant species complex, namely the Louisiana Irises, into a ‘model system’ for investigating hybrid fitness and the role of genetic exchange in adaptive evolution and diversification. In particular, we will argue that a multitude of approaches, involving both experimental and natural environments, and incorporating both manipulative analyses and surveys of natural populations, are necessary to adequately test for the evolutionary significance of introgressive hybridization. An appreciation of the variability of hybrid fitness leads to the conclusion that certain genetic signatures reflect adaptive evolution. Furthermore, tests of the frequency of allopatric versus sympatric/parapatric divergence (that is, divergence with ongoing gene flow) support hybrid genotypes as a mechanism of evolutionary diversification in numerous species complexes. -

Lepidoptera, Incurvariidae) with Two New Species from China and Japan
Zootaxa 4927 (2): 209–233 ISSN 1175-5326 (print edition) https://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2021 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4927.2.3 http://zoobank.org/urn:lsid:zoobank.org:pub:96B9981B-01B5-4828-A4C6-E2E4A08DB8F2 Review of the genus Vespina (Lepidoptera, Incurvariidae) with two new species from China and Japan TOSHIYA HIROWATARI1*, SADAHISA YAGI1, ISSEI OHSHIMA2, GUO-HUA HUANG3 & MIN WANG4 1Entomological laboratory, Faculty of Agriculture, Kyushu University, Fukuoka, 819-0395 Japan. [email protected]; https://orcid.org/0000-0002-4261-1219 2Department of Life and Environmental Sciences, Kyoto Prefectural University, Kyoto, 606-8522 Japan. [email protected]; https://orcid.org/0000-0001-8295-9749 3Hunan Provincial Key Laboratory for Biology and Control of Plant Diseases and Insect Pests, Hunan Agricultural University, Changsha 410128, Hunan, China. [email protected]; https://orcid.org/0000-0002-6841-0095 4Department of Entomology, South China Agricultural University, Guangzhou 510640, Guangdong, China. [email protected]; https://orcid.org/0000-0001-5834-4058 *Corresponding author. [email protected]; https://orcid.org/0000-0002-6839-2229 Abstract Asian species of the genus Vespina Davis, 1972 (Lepidoptera, Incurvariidae) are mainly reviewed. Vespina meridiana Hirowatari & Yagi sp. nov. from the Ryukyu Islands, Japan, and Vespina sichuana Hirowatari, Huang & Wang sp. nov. from Sichuan, China, are described. The previously known Vespina species are associated with plants from the Fagaceae family on the western coast of the USA and East Asia and with Sapindaceae (Aceraceae) in eastern Europe. -
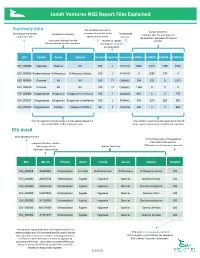
NGS Report Explained
Jonah Ventures NGS Report Files Explained Summary data The % of base pairs in the Sample identifiers Exact Sequence Variant sequence that match to the The detected Exact Sequence Variant Consensus Taxonomy The detected Numbers after the decimal point unique identifier species in the library. sequence. represent lab replicates of the same Taxonomic ranking from the Number of species sample. library matched to each sequence. matching the sequence at a given level. ESV Family Genus Species %match # Species Sequence S10001.1 S10002.1 S10003.1 S10003.2 ESV_000080 Fagaceae Quercus NA 100 7 AAAAAG… 3066 1340 5780 3462 ESV_000818 Orobanchaceae Orthocarpus Orthocarpus luteus 100 1 AAAAAG… 0 2582 249 0 ESV_000006 Poaceae NA NA 100 77 GAAAAG… 584 101 0 1039 ESV_000050 Poaceae NA NA 100 12 GAAAAG… 1328 0 0 0 ESV_000308 Polygonaceae Polygonum Polygonum humifusum 100 2 AAAAAG… 902 0 0 278 ESV_000027 Polygonaceae Eriogonum Eriogonum umbellatum 100 1 ATAAAG… 341 124 282 381 ESV_000520 Polygonaceae Fallopia Fallopia multiflora 99 1 AAAAAG… 126 0 0 994 “NA” at a taxonomic ranks means multiple species present in The number in each cell is the absolute number of the sample differ in that taxonomic rank times a given sequence was read by the sequencer. ESV detail Exact Sequence Variant The % of base pairs in the sequence that match to the species. Index identification number. that match to the species. Each sample has an Species Taxonomy 100% indicates all base pairs matched individual index number. ESV GB_ID Phylum Order Family Genus Species %match ESV_000818 -

Ray Imaging of a Dichasium Cupule of Castanopsis from Eocene Baltic Amber
RESEARCH ARTICLE Synchrotron X- ray imaging of a dichasium cupule of Castanopsis from Eocene Baltic amber Eva-Maria Sadowski1,4 , Jörg U. Hammel2 , and Thomas Denk3 Manuscript received 30 May 2018; revision accepted 6 September PREMISE OF THE STUDY: The Eocene Baltic amber deposit represents the largest 2018. accumulation of fossil resin worldwide, and hundreds of thousands of entrapped 1 Department of Geobiology, University of Göttingen, arthropods have been recovered. Although Baltic amber preserves delicate plant Goldschmidtstraße 3, 37077 Göttingen, Germany structures in high fidelity, angiosperms of the “Baltic amber forest” remain poorly studied. 2 Institute of Materials Research, Helmholtz-Zentrum Geesthacht, We describe a pistillate partial inflorescence of Castanopsis (Fagaceae), expanding the Max-Planck-Str. 1, 21502 Geesthacht, Germany knowledge of Fagaceae diversity from Baltic amber. 3 Department of Palaeobiology, Swedish Museum of Natural History, Box 50007, 10405 Stockholm, Sweden METHODS: The amber specimen was investigated using light microscopy and 4 Author for correspondence (e-mail: eva-maria.sadowski@ synchrotron- radiation- based X- ray micro- computed tomography (SRμCT). geo.uni-goettingen.de) KEY RESULTS: The partial inflorescence is a cymule, consisting of an involucre of scales Citation: Sadowski, E.-M., J. U. Hammel, and T. Denk. 2018. Synchrotron X- ray imaging of a dichasium cupule of Castanopsis that surround all four pistillate flowers, indicating a dichasium cupule. Subtending bracts from Eocene Baltic amber. American Journal of Botany 105(12): are basally covered with peltate trichomes. Flowers possess an urecolate perianth of 2025–2036. six nearly free lobes, 12 staminodia hidden by the perianth, and a tri-locular ovary that doi:10.1002/ajb2.1202 is convex- triangular in cross section.