Inhibitory Effects of Midostaurin and Avapritinib on Myeloid Progenitors Derived from Patients with KIT D816V Positive Advanced Systemic Mastocytosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NCCN Guidelines for Myeloid/Lymphoid Neoplasms with Eosinophilia and Tyrosine Kinase Fusion Genes V.2.2021 –Interim on 07/21/20
NCCN Guidelines for Myeloid/Lymphoid Neoplasms with Eosinophilia and Tyrosine Kinase Fusion Genes V.2.2021 –Interim on 07/21/20 Guideline Page Institution Vote Panel Discussion/References and Request YES NO ABSTAIN ABSENT MLNE-5 The panel consensus was to include the use of avapritinib in a 18 1 1 6 Internal request: clinical trial with the following footnote: "Avapritinib is approved for adult patients with unresectable or metastatic Consider the addition of avapritinib for gastrointestinal stromal tumors (GISTs) harboring a PDGFRA Myeloid/lymphoid Neoplasms with FIP1L1- exon 18 mutation, including D842V mutations. This suggests a PDGFRA rearrangement in patients with possible role for avapritinib in patients with FIP1L1-PDGFRA– PDGFRA D842V mutation. positive myeloid/lymphoid neoplasms with eosinophilia harboring PDGFRA D842V mutation resistant to imatinib. If this mutation is identified, a clinical trial of avapritinib is preferred (if available), rather than off-label use." MLNE-7 The panel consensus was to include pemigatinib as a TKI with 18 1 1 6 Internal request: activity against FGFR1 under other recommended regimens with the following footnote corresponding to clinical trial and Consider the addition of the pemigatinib for pemigatinib: "Pemigatinib (FGFR1, 2, and 3 inhibitor) is Myeloid/lymphoid Neoplasms with FGFR1 approved for the treatment of adult patients with previously rearrangement. treated, unresectable, locally advanced or metastatic cholangiocarcinoma and a FGFR2 fusion or other rearrangement, as detected by an FDA-approved test. Pemigatinib has received orphan drug designation for the treatment of patients with myeloid/lymphoid neoplasms with eosinophilia and FGFR1 rearrangement and is currently being evaluated in a clinical trial for this indication. -

Leukemia Insights June 2021
JUNE 2021 In this month’s Leukemia Insights newsletter, written by Prithviraj Bose, M.D. and Srdan Verstovsek, M.D., Ph.D., and sponsored in part by the Charif Souki Cancer Research ABOUT MyMDAnderson Fund, we discuss our novel therapeutic approaches for the rare hematologic malignancies, systemic mastocytosis and myeloid/lymphoid neoplasms with myMDAnderson is a secure, eosinophilia. Learn more about our Leukemia program. personalized web site helping community physicians expedite patient referrals, as well as improve continuity Spotlight on rare, atypical, myeloid of care through information access and streamlined communications. neoplasms: systemic mastocytosis and Physicians who have referred patients myeloid/lymphoid neoplasms with to MD Anderson or plan to do so, can utilize the HIPAA compliant features eosinophilia and FGFR1 rearrangements of myMDAnderson to: • Refer a patient • View your patient's appointments Systemic Mastocytosis Access patient reports • Send and receive secure Systemic mastocytosis (SM) is a rare myeloid neoplasm messages driven in approximately 95% of cases by an activating mutation in c-KIT, usually D816V. SM is characterized as indolent, smoldering or advanced, based on the presence JOIN THE COVERSATION and number of so-called B- and C-findings. The latter Connect with us. signify organ damage and are a hallmark of advanced SM (AdvSM). AdvSM, in turn, is typically sub-classified as aggressive SM (ASM), SM with an associated hematologic neoplasm (SM-AHN, the most common subtype) and mast cell leukemia (MCL). Patients with indolent SM (ISM) and smoldering SM (SSM) enjoy much better survival than JOIN OUR MAILING LIST those with AdvSM, although symptoms in all three subtypes can be severe and debilitating. -
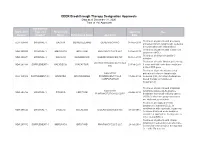
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

PIONEER: a Randomized, Double-Blind, Placebo-Controlled, Phase 2 Study of Avapritinib in Patients with Indolent Or Smoldering Sy
PIONEER: A Randomized, Double-Blind, Placebo-Controlled, Phase 2 Study of Avapritinib in Patients with Indolent or Smoldering Systemic Mastocytosis with Symptoms Inadequately Controlled with Standard Therapy Cem Akin, Hanneke Oude Elberink, Jason Gotlib, Vito Sabato, Karin Hartmann, Sigurd Broesby-Olsen, Mariana Castells, Michael W. Deininger, Mark L. Heaney, Tracy I. George, Deepti Radia, Massimo Triggiani, Paul van Daele, Daniel J. DeAngelo, Oleg Schmidt-Kittler, Hui-Min Lin, Andrew Morrison, Brenton Mar, Frank Siebenhaar, Marcus Maurer American Academy of Allergy Asthma and Immunology Annual Meeting March 16, 2020 Disclosures • Investigator: Blueprint Medicines’ ongoing Phase 2 PIONEER trial in indolent and smoldering systemic mastocytosis • Consultant: Blueprint Medicines, Novartis • AYVAKIT™ (avapritinib) is approved by the FDA for the treatment of adults with unresectable or metastatic gastrointestinal stromal tumor (GIST) harboring a platelet-derived growth factor receptor alpha (PDGFRA) exon 18 mutation, including PDGFRA D842V mutations, in the United States. Avapritinib has not been approved by the FDA or any other health authority for use in the United States for any other indication or in any other jurisdiction for any indication. • All data in this presentation are based on a cut-off date of December 27, 2019 unless otherwise specified. 2 Systemic mastocytosis (SM) is a clonal mast cell (MC) neoplasm driven by KIT D816V SM Prevalence of ~1:10,000 ~32,000 estimated in US KIT D816V ~5% Advanced SM Organ damage and decreased survival -

Leukemia Insights Newsletter September 2019 MPN
SEPTEMBER)2019 In this month’s Leukemia Insights newsletter, written by Prithviraj Bose, MD, and Srdan Verstovsek, PhD, MD, and sponsoreD in part by the Charif Souki Cancer Research ABOUT)MyMDAnderson FunD, we summarize the investigational approaches available for patients with myeloproliferative neoplasms (MPN) at MD AnDerson Cancer Center. myMDAnderson is`a`seCure,` personalized`web`site`helping` community`physicians`expedite`patient` referrals,`as`well`as`improve`Continuity` Innovative)Treatment)Strategies)for) of`Care`through`information`acCess` Classic)and)Atypical)Myeloproliferative) and`streamlined`CommuniCations.` PhysiCians`who`have`referred`patients` Neoplasms)(MPN) to`MD`Anderson`or`plan`to`do`so,`Can` utilize`the`HIPAA`Compliant`features In August 2019, the FDA approved the JAK2 inhibitor fedratinib for the of`myMDAnderson`to: treatment of patients with myelofibrosis (MF), only the second drug approval in the classiC myeloproliferative neoplasm (MPN) space after • Refer`a`patient ruxolitinib was approved for MF in 2011 and polyCythemia vera (PV) in • View`your`patient's`appointments` 2014. Hopefully, this is the first of many regulatory approvals as a AcCess`patient`reports number of investigational agents appear promising in reCent and • Send`and`reCeive`seCure` ongoing trials. Below, we summarize the investigational approaches messages available for patients with MPN at MD Anderson. 1.)Pemigatinib for)myeloid/lymphoid)neoplasms)with)FGFR1 JOIN)THE)COVERSATION rearrangement)(NCT03011372) Connect+with+us. Myeloid/lymphoid neoplasms with a rearrangement of the FGFR1 gene loCated at 8p11.2 represent an exCeedingly rare but aggressive malignanCy with no standard treatment options and a dismal prognosis. Eosinophilia is usually present and should trigger testing for rearrangements involving FGFR1, and also PDGFRα and PDGFRβ. -

GIST): Efficacy and Safety Data from Phase 3 VOYAGER Study 3461883 Yoon-Koo Kang1, Suzanne George2, Robin L
Avapritinib vs Regorafenib in Patients With Locally Advanced Unresectable or Metastatic Gastrointestinal Stromal Tumor Abstract (GIST): Efficacy and Safety Data From Phase 3 VOYAGER Study 3461883 Yoon-Koo Kang1, Suzanne George2, Robin L. Jones3, Piotr Rutkowski4, Lin Shen5, Olivier Mir6, Shreyaskumar Patel7, Yongjian Zhou8, Margaret von Mehren9, Peter Hohenberger10, Victor Villalobos11, Mehdi Brahmi12, William D. Tap13, Jonathan Trent14, Maria A. Pantaleo15, Patrick Schöffski16, Kevin He17, Paggy Hew17, Kate Newberry17, Maria Roche17, Michael Heinrich18, Sebastian Bauer19 1Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea; 2Sarcoma Center, Dana Farber Cancer Institute, Boston, MA, USA; 3Royal Marsden Hospital and Institute of Cancer Research, London, UK; 4Maria Sklodowska-Curie National Research Institute of Oncology, Warsaw, Poland; 5Peking University Cancer Hospital and Institute, Beijing, China; 6Institut Gustave Roussy, Villejuif, France; 7MD Anderson Cancer Center, Houston, TX, USA; 8Fujian Medical University Union Hospital, Fuzhou, China; 9Fox Chase Cancer Center, Philadelphia, PA, USA; 10Medical Faculty Mannheim (UMM), University of Heidelberg, Mannheim, Germany; 11University of Colorado Denver – Anschutz Medical Campus, Aurora, CO, USA; 12Centre Léon Bérard, Lyon, France; 13Memorial Sloan Kettering Cancer Center and Weill Cornell Medical College, New York, NY, USA; 14University of Miami-Sylvester Comprehensive Cancer Center, Miami, FL, USA; 15University of Bologna, Bologna, Italy; 16University Hospitals -

AYVAKIT (Avapritinib) RATIONALE for INCLUSION in PA PROGRAM
AYVAKIT (avapritinib) RATIONALE FOR INCLUSION IN PA PROGRAM Background Ayvakit (avapritinib) is a tyrosine kinase inhibitor that targets platelet-derived growth factor receptor alpha (PDGFRA) and PDGFRA D842 mutants as well as multiple KIT exon 11, 11/17 and 17 mutants. Certain mutations in PDGFRA and KIT can result in the autophosphorylation and constitutive activation of these receptors which can contribute to tumor cell proliferation. Other potential targets for Ayvakit include wild type KIT, PDGFRB, and CSFR1. Ayvakit inhibits the autophosphorylation of KIT D816V and PDGFRA D842V, mutants associated with resistance to approved kinase inhibitors. This could contribute to its inhibition of tumor cell proliferation (1). Regulatory Status FDA-approved indications: Ayvakit is a kinase inhibitor indicated for: (1) • Gastrointestinal Stromal Tumor (GIST) o the treatment of adults with unresectable or metastatic GIST harboring a platelet- derived growth factor receptor alpha (PDGFRA) exon 18 mutation, including PDGFRA D842V mutations • Advanced Systemic Mastocyosis (AdvSM) o the treatment of adult patients with AdvSM. AdvSM includes patients with aggressive systemic mastocytosis (ASM), systemic mastocytosis with an associated hematological neoplasm (SM-AHN), and mast cell leukemia (MCL). o Limitations of Use: Ayvakit is not recommended for the treatment of patients with AdvSM with platelet counts of less than 50 x 109/L Ayvakit has warnings regarding intracranial hemorrhage, cognitive effects and embryo-fetal toxicity. Serious intracranial hemorrhage may occur in patients treated with Ayvakit. Ayvakit should be discontinued if intracranial hemorrhage of any grade occurs. A broad spectrum of cognitive adverse reactions can occur in patients receiving Ayvakit. Ayvakit can cause fetal harm when administered to pregnant women. -

Tyrosine Kinase Inhibitors for Solid Tumors in the Past 20 Years (2001–2020) Liling Huang†, Shiyu Jiang† and Yuankai Shi*
Huang et al. J Hematol Oncol (2020) 13:143 https://doi.org/10.1186/s13045-020-00977-0 REVIEW Open Access Tyrosine kinase inhibitors for solid tumors in the past 20 years (2001–2020) Liling Huang†, Shiyu Jiang† and Yuankai Shi* Abstract Tyrosine kinases are implicated in tumorigenesis and progression, and have emerged as major targets for drug discovery. Tyrosine kinase inhibitors (TKIs) inhibit corresponding kinases from phosphorylating tyrosine residues of their substrates and then block the activation of downstream signaling pathways. Over the past 20 years, multiple robust and well-tolerated TKIs with single or multiple targets including EGFR, ALK, ROS1, HER2, NTRK, VEGFR, RET, MET, MEK, FGFR, PDGFR, and KIT have been developed, contributing to the realization of precision cancer medicine based on individual patient’s genetic alteration features. TKIs have dramatically improved patients’ survival and quality of life, and shifted treatment paradigm of various solid tumors. In this article, we summarized the developing history of TKIs for treatment of solid tumors, aiming to provide up-to-date evidence for clinical decision-making and insight for future studies. Keywords: Tyrosine kinase inhibitors, Solid tumors, Targeted therapy Introduction activation of tyrosine kinases due to mutations, translo- According to GLOBOCAN 2018, an estimated 18.1 cations, or amplifcations is implicated in tumorigenesis, million new cancer cases and 9.6 million cancer deaths progression, invasion, and metastasis of malignancies. occurred in 2018 worldwide [1]. Targeted agents are In addition, wild-type tyrosine kinases can also func- superior to traditional chemotherapeutic ones in selec- tion as critical nodes for pathway activation in cancer. -

ATP-Competitive Inhibitors Midostaurin and Avapritinib Have Distinct Resistance Profiles in Exon 17–Mutant KIT Beth Apsel Winger1, Wilian A
Published OnlineFirst July 3, 2019; DOI: 10.1158/0008-5472.CAN-18-3139 Cancer Translational Science Research ATP-Competitive Inhibitors Midostaurin and Avapritinib Have Distinct Resistance Profiles in Exon 17–Mutant KIT Beth Apsel Winger1, Wilian A. Cortopassi2, Diego Garrido Ruiz2, Lucky Ding3, Kibeom Jang3, Ariel Leyte-Vidal3, Na Zhang2,4, Rosaura Esteve-Puig5, Matthew P. Jacobson2, and Neil P. Shah3 Abstract KIT is a type-3 receptor tyrosine kinase that is frequently domain substitutions, with the T670I gatekeeper mutation mutated at exon 11 or 17 in a variety of cancers. First- being selectively problematic for avapritinib. Although generation KIT tyrosine kinase inhibitors (TKI) are ineffec- gatekeeper mutations often directly disrupt inhibitor bind- tive against KIT exon 17 mutations, which favor an active ing, we provide evidence that T670I confers avapritinib conformation that prevents these TKIs from binding. resistance indirectly by inducing distant conformational The ATP-competitive inhibitors, midostaurin and avapriti- changes in the phosphate-binding loop. These findings nib, which target the active kinase conformation, were suggest combining midostaurin and avapritinib may fore- developed to inhibit exon 17–mutant KIT. Because second- stall acquired resistance mediated by secondary kinase ary kinase domain mutations are a common mechanism domain mutations. of TKI resistance and guide ensuing TKI design, we sought to define problematic KIT kinase domain mutations for Significance:Thisstudyidentifies potential problematic these emerging therapeutics. Midostaurin and avapritinib kinase domain mutations for next-generation KIT inhibitors displayed different vulnerabilities to secondary kinase midostaurin and avapritinib. Introduction 17, which encodes the activation loop of the kinase domain (2–5). Exon 11 mutations activate KIT by relieving the autoinhibition KIT is a type-3 receptor tyrosine kinase (RTK); other type-3 RTKs of the JM domain, while exon 17 mutations shift the conforma- are FLT3, PDGFR, and CSF1R. -

Revolutions in Treatment Options in Gastrointestinal Stromal Tumours (Gists): the Latest Updates Revolutions in Treatment Options in GIST
Curr. Treat. Options in Oncol. (2020) 21:55 DOI 10.1007/s11864-020-00754-8 Sarcoma (SH Okuno, Section Editor) Revolutions in treatment options in gastrointestinal stromal tumours (GISTs): the latest updates Revolutions in treatment options in GIST Sheima Farag, MD1 Myles J. Smith, MB, BCh, BAO, PhD, FRCSI, FRCS1 Nicos Fotiadis, MD, PhD, FRCR1 Anastasia Constantinidou, MD, MSc, MRCP, PhD2 Robin L. Jones, BSc, MB, BS, MRCP, MD(Res)1,* Address *,1Sarcoma Unit, The Royal Marsden Hospital (NHS Foundation Trust) and Institute of Cancer Research, Fulham Road, London, SW3 6JJ, UK Email: [email protected] 2Medical School University of Cyprus and BoC Oncology Centre, Nicosia, Cyprus * The Author(s) 2020 Anastasia Constantinidou and Robin L. Jones contributed equally to this work. This article is part of the Topical Collection on Sarcoma Keywords Gastrointestinal stromal tumour I GIST I Tyrosine kinase inhibitors I TKI I Avapritinib I Ripretinib Opinion statement The treatment of advanced GIST is rapidly evolving with the development of novel molecular compounds such as avapritinib and ripretinib, but also promising results have been achieved with cabozantinib in a phase II trial. The availability of over five lines of treatment for patients with advanced GIST is likely to completely shift the current second-line and third-line treatment options, and will also potentially enable a personalised approach to treatment. Imatinib will most likely remain as the first-line treatment of choice for the vast majority of GIST patients. However, for GIST patients with tumours harbouring a D842V mutation in PDGFRA exon 18, avapritinib has shown efficacy and will become first-line therapy for this molecular subgroup. -

Multi-Discipline Review
CENTER FOR DRUG EVALUATION AND RESEARCH APPLICATION NUMBER: 212608Orig1s000 MULTI-DISCIPLINE REVIEW Summary Review Office Director Cross Discipline Team Leader Review Clinical Review Non-Clinical Review Statistical Review Clinical Pharmacology Review NDA/BLA Multi-disciplinary Review and Evaluation NDA 212608 AYVAKIT (avapritinib) NDA/BLA Multi-Disciplinary Review and Evaluation Application Type New Drug Application (NDA)/New Molecular Entity (NME) Application Number(s) 212608 Priority or Standard Priority Submit Date(s) June 14, 2019 Received Date(s) June 14, 2019 PDUFA Goal Date February 14, 2020 Division/Office DO3/OOD Established/Proper Name Avapritinib (Proposed) Trade Name AYVAKIT Pharmacologic Class Kinase inhibitor Code name BLU-285 Applicant Blueprint Medicines Corporation Dosage form 100 mg, 200 mg, and 300 mg tablets Applicant proposed Dosing 300 mg orally once daily Regimen Applicant Proposed For the treatment of adult patients with unresectable or Indication(s)/Population(s) metastatic gastrointestinal stromal tumor (GIST) who have a platelet-derived growth factor receptor alpha (PDGFA) exon 18 mutation, regardless of prior therapy Applicant Proposed SNOMED CT Indication Gastrointestinal Stromal Tumor Disease Term for each Proposed Indication Recommendation on Approval Regulatory Action Recommended For the treatment of adults with unresectable or metastatic Indication(s)/Population(s) GIST harboring a platelet-derived growth factor receptor alpha (if applicable) (PDGFRA) exon 18 mutation, including PDGFRA D842V mutations Recommended SNOMED CT Indication Disease Gastrointestinal Stromal Tumor Term for each Indication (if applicable) Recommended Dosing 300 mg orally once daily Regimen 1 Version date: April 2, 2018 Reference ID: 4543562 NDA/BLA Multi-disciplinary Review and Evaluation NDA 212608 AYVAKIT (avapritinib) Table of Contents Table of Tables ............................................................................................................................... -

Oncology Approvals in 2020: a Year of Firsts in the Midst of a Pandemic
COMMENT Oncology approvals in 2020: a year of firsts in the midst of a pandemic Laleh Amiri- Kordestani1 ✉ and Richard Pazdur1,2 In 2020, despite challenges related to the COVID-19 pandemic, the US FDA approved 30 new drugs and biologic agents, 45 supplemental drug and biologic applications and 1 biosimilar application in oncology. To limit potential exposure to SARS- CoV-2, in 2020 approved for first- line treatment of patients with meta- the FDA the FDA made several approvals aimed at reducing the static microsatellite instability- high or mismatch made several frequency of interactions between patients with cancer repair- deficient colorectal cancer, and brexucabtagene approvals aimed and their health-care providers (Supplementary Table 1). autoleucel became the first chimeric antigen receptor These approaches included increasing dosing intervals, T cell therapy approved for mantle cell lymphoma. at reducing the such as that for pembrolizumab (which was extended In 2020, tucatinib, in combination with trastu- frequency of from 3 weeks to 6 weeks), and adopting oral or subcu- zumab and capecitabine for patients with metastatic interactions taneous formulations of already approved drugs. New HER2- positive breast cancer (including those with brain between patients formulations approved in 2020 included three different metastases), was the first new molecular entity reviewed intravenous dosing regimens of atezolizumab, the first under the Oncology Center of Excellence (OCE) Project with cancer and oral tablet combining decitabine plus cedazuridine, Orbis1. This project provides a framework for concurrent their health-care a sub cutaneously injectable combination of dara- submission and review of oncology drugs among interna- providers tumumab plus hyaluronidase- fihj, and a fixed- dose tional partners, with the goal of providing patients faster combination of pertuzumab, trastuzumab and hyalu- access to drugs in countries in which substantial delays ronidase–zzxf.