ATP-Competitive Inhibitors Midostaurin and Avapritinib Have Distinct Resistance Profiles in Exon 17–Mutant KIT Beth Apsel Winger1, Wilian A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FLT3 Inhibitors in Acute Myeloid Leukemia Mei Wu1, Chuntuan Li2 and Xiongpeng Zhu2*
Wu et al. Journal of Hematology & Oncology (2018) 11:133 https://doi.org/10.1186/s13045-018-0675-4 REVIEW Open Access FLT3 inhibitors in acute myeloid leukemia Mei Wu1, Chuntuan Li2 and Xiongpeng Zhu2* Abstract FLT3 mutations are one of the most common findings in acute myeloid leukemia (AML). FLT3 inhibitors have been in active clinical development. Midostaurin as the first-in-class FLT3 inhibitor has been approved for treatment of patients with FLT3-mutated AML. In this review, we summarized the preclinical and clinical studies on new FLT3 inhibitors, including sorafenib, lestaurtinib, sunitinib, tandutinib, quizartinib, midostaurin, gilteritinib, crenolanib, cabozantinib, Sel24-B489, G-749, AMG 925, TTT-3002, and FF-10101. New generation FLT3 inhibitors and combination therapies may overcome resistance to first-generation agents. Keywords: FMS-like tyrosine kinase 3 inhibitors, Acute myeloid leukemia, Midostaurin, FLT3 Introduction RAS, MEK, and PI3K/AKT pathways [10], and ultim- Acute myeloid leukemia (AML) remains a highly resist- ately causes suppression of apoptosis and differentiation ant disease to conventional chemotherapy, with a me- of leukemic cells, including dysregulation of leukemic dian survival of only 4 months for relapsed and/or cell proliferation [11]. refractory disease [1]. Molecular profiling by PCR and Multiple FLT3 inhibitors are in clinical trials for treat- next-generation sequencing has revealed a variety of re- ing patients with FLT3/ITD-mutated AML. In this re- current gene mutations [2–4]. New agents are rapidly view, we summarized the preclinical and clinical studies emerging as targeted therapy for high-risk AML [5, 6]. on new FLT3 inhibitors, including sorafenib, lestaurtinib, In 1996, FMS-like tyrosine kinase 3/internal tandem du- sunitinib, tandutinib, quizartinib, midostaurin, gilteriti- plication (FLT3/ITD) was first recognized as a frequently nib, crenolanib, cabozantinib, Sel24-B489, G-749, AMG mutated gene in AML [7]. -

Federal Register Notice 5-1-2020 Pdf Icon[PDF – 358
Federal Register / Vol. 85, No. 85 / Friday, May 1, 2020 / Notices 25439 confidential by the respondent (5 U.S.C. schedules. Other than examination DEPARTMENT OF HEALTH AND 552(b)(4)). reports, it provides the only financial HUMAN SERVICES Current actions: The Board has data available for these corporations. temporarily revised the instructions to The Federal Reserve is solely Centers for Disease Control and the FR Y–9C report to accurately reflect responsible for authorizing, supervising, Prevention the revised definition of ‘‘savings and assigning ratings to Edges. The [CDC–2020–0046; NIOSH–233–C] deposits’’ in accordance with the Federal Reserve uses the data collected amendments to Regulation D in the on the FR 2886b to identify present and Hazardous Drugs: Draft NIOSH List of interim final rule published on April 28, potential problems and monitor and Hazardous Drugs in Healthcare 2020 (85 FR 23445). Specifically, the develop a better understanding of Settings, 2020; Procedures; and Risk Board has temporarily revised the activities within the industry. Management Information instructions on the FR Y–9C, Schedule HC–E, items 1(b), 1(c), 2(c) and glossary Legal authorization and AGENCY: Centers for Disease Control and content to remove the transfer or confidentiality: Sections 25 and 25A of Prevention, HHS. withdrawal limit. As a result of the the Federal Reserve Act authorize the ACTION: Notice and request for comment. revision, if a depository institution Federal Reserve to collect the FR 2886b chooses to suspend enforcement of the (12 U.S.C. 602, 625). The obligation to SUMMARY: The National Institute for six transfer limit on a ‘‘savings deposit,’’ report this information is mandatory. -

Efficacy and Safety of Midostaurin-Based Induction and Maintenance Therapy for Newly Diagnosed AML
POST-ASH Issue 4, 2016 Efficacy and Safety of Midostaurin-Based Induction and Maintenance Therapy for Newly Diagnosed AML For more visit ResearchToPractice.com/5MJCASH2016 CME INFORMATION OVERVIEW OF ACTIVITY Each year, thousands of clinicians, basic scientists and other industry professionals sojourn to major international oncology conferences, like the American Society of Hematology (ASH) annual meeting, to hone their skills, network with colleagues and learn about recent advances altering state-of-the-art management in hematologic oncology. These events have become global stages where exciting science, cutting-edge concepts and practice-changing data emerge on a truly grand scale. This massive outpouring of information has enormous benefits for the hematologic oncology community, but the truth is it also creates a major challenge for practicing oncologists and hematologists. Although original data are consistently being presented and published, the flood of information unveiled during a major academic conference is unmatched and leaves in its wake an enormous volume of new knowledge that practicing oncologists must try to sift through, evaluate and consider applying. Unfortunately and quite commonly, time constraints and an inability to access these data sets leave many oncologists struggling to ensure that they’re aware of crucial practice-altering findings. This creates an almost insurmountable obstacle for clinicians in community practice because they are not only confronted almost overnight with thousands of new presentations and -

NCCN Guidelines for Myeloid/Lymphoid Neoplasms with Eosinophilia and Tyrosine Kinase Fusion Genes V.2.2021 –Interim on 07/21/20
NCCN Guidelines for Myeloid/Lymphoid Neoplasms with Eosinophilia and Tyrosine Kinase Fusion Genes V.2.2021 –Interim on 07/21/20 Guideline Page Institution Vote Panel Discussion/References and Request YES NO ABSTAIN ABSENT MLNE-5 The panel consensus was to include the use of avapritinib in a 18 1 1 6 Internal request: clinical trial with the following footnote: "Avapritinib is approved for adult patients with unresectable or metastatic Consider the addition of avapritinib for gastrointestinal stromal tumors (GISTs) harboring a PDGFRA Myeloid/lymphoid Neoplasms with FIP1L1- exon 18 mutation, including D842V mutations. This suggests a PDGFRA rearrangement in patients with possible role for avapritinib in patients with FIP1L1-PDGFRA– PDGFRA D842V mutation. positive myeloid/lymphoid neoplasms with eosinophilia harboring PDGFRA D842V mutation resistant to imatinib. If this mutation is identified, a clinical trial of avapritinib is preferred (if available), rather than off-label use." MLNE-7 The panel consensus was to include pemigatinib as a TKI with 18 1 1 6 Internal request: activity against FGFR1 under other recommended regimens with the following footnote corresponding to clinical trial and Consider the addition of the pemigatinib for pemigatinib: "Pemigatinib (FGFR1, 2, and 3 inhibitor) is Myeloid/lymphoid Neoplasms with FGFR1 approved for the treatment of adult patients with previously rearrangement. treated, unresectable, locally advanced or metastatic cholangiocarcinoma and a FGFR2 fusion or other rearrangement, as detected by an FDA-approved test. Pemigatinib has received orphan drug designation for the treatment of patients with myeloid/lymphoid neoplasms with eosinophilia and FGFR1 rearrangement and is currently being evaluated in a clinical trial for this indication. -

Disruption of CSF-1R Signaling Inhibits Growth of AML with Inv(16)
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2021 Disruption of CSF-1R signaling inhibits growth of AML with inv(16) Simonis, Alexander ; Russkamp, Norman F ; Mueller, Jan ; Wilk, C Matthias ; Wildschut, Mattheus H E ; Myburgh, Renier ; Wildner-Verhey van Wijk, Nicole ; Mueller, Rouven ; Balabanov, Stefan ; Valk, Peter J M ; Theocharides, Alexandre P A ; Manz, Markus G DOI: https://doi.org/10.1182/bloodadvances.2020003125 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-202789 Journal Article Published Version The following work is licensed under a Publisher License. Originally published at: Simonis, Alexander; Russkamp, Norman F; Mueller, Jan; Wilk, C Matthias; Wildschut, Mattheus H E; Myburgh, Renier; Wildner-Verhey van Wijk, Nicole; Mueller, Rouven; Balabanov, Stefan; Valk, Peter J M; Theocharides, Alexandre P A; Manz, Markus G (2021). Disruption of CSF-1R signaling inhibits growth of AML with inv(16). Blood advances, 5(5):1273-1277. DOI: https://doi.org/10.1182/bloodadvances.2020003125 STIMULUS REPORT Disruption of CSF-1R signaling inhibits growth of AML with inv(16) Alexander Simonis,1,* Norman F. Russkamp,1,* Jan Mueller,1 C. Matthias Wilk,1 Mattheus H. E. Wildschut,1,2 Renier Myburgh,1 Nicole Wildner-Verhey van Wijk,1 Rouven Mueller,1 Stefan Balabanov,1 Peter J. M. Valk,3 Alexandre P. A. Theocharides,1 and Markus G. Manz1 1Department of Medical Oncology and Hematology, University Hospital -

Leukemia Insights June 2021
JUNE 2021 In this month’s Leukemia Insights newsletter, written by Prithviraj Bose, M.D. and Srdan Verstovsek, M.D., Ph.D., and sponsored in part by the Charif Souki Cancer Research ABOUT MyMDAnderson Fund, we discuss our novel therapeutic approaches for the rare hematologic malignancies, systemic mastocytosis and myeloid/lymphoid neoplasms with myMDAnderson is a secure, eosinophilia. Learn more about our Leukemia program. personalized web site helping community physicians expedite patient referrals, as well as improve continuity Spotlight on rare, atypical, myeloid of care through information access and streamlined communications. neoplasms: systemic mastocytosis and Physicians who have referred patients myeloid/lymphoid neoplasms with to MD Anderson or plan to do so, can utilize the HIPAA compliant features eosinophilia and FGFR1 rearrangements of myMDAnderson to: • Refer a patient • View your patient's appointments Systemic Mastocytosis Access patient reports • Send and receive secure Systemic mastocytosis (SM) is a rare myeloid neoplasm messages driven in approximately 95% of cases by an activating mutation in c-KIT, usually D816V. SM is characterized as indolent, smoldering or advanced, based on the presence JOIN THE COVERSATION and number of so-called B- and C-findings. The latter Connect with us. signify organ damage and are a hallmark of advanced SM (AdvSM). AdvSM, in turn, is typically sub-classified as aggressive SM (ASM), SM with an associated hematologic neoplasm (SM-AHN, the most common subtype) and mast cell leukemia (MCL). Patients with indolent SM (ISM) and smoldering SM (SSM) enjoy much better survival than JOIN OUR MAILING LIST those with AdvSM, although symptoms in all three subtypes can be severe and debilitating. -
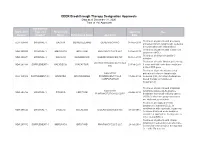
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

Recommendations from York and Scarborough Medicines
Recommendations from York and Scarborough Medicines Commissioning Committee July 2018 Drug name Indication Recommendation, rationale and place in RAG status Potential full year cost impact therapy CCG commissioned Technology Appraisals 1. Nil NHSE commissioned Technology Appraisals – for noting 2. TA520: Atezolizumab for Atezolizumab is recommended as an option for Red No cost impact to CCGs as NHS England treating locally advanced or treating locally advanced or metastatic non- commissioned. metastatic non-small-cell lung small-cell lung cancer (NSCLC) in adults who cancer after chemotherapy have had chemotherapy (and targeted treatment if they have an EGFR- or ALK‑ positive tumour), only if: atezolizumab is stopped at 2 years of uninterrupted treatment or earlier if the disease progresses and the company provides atezolizumab with the discount agreed in the patient access scheme. 3. TA522: Pembrolizumab for Pembrolizumab is recommended for use within Red No cost impact to CCGs as NHS England untreated locally advanced or the Cancer Drugs Fund as an option for commissioned. metastatic urothelial cancer untreated locally advanced or metastatic when cisplatin is unsuitable urothelial carcinoma in adults when cisplatin- containing chemotherapy is unsuitable, only if: pembrolizumab is stopped at 2 years of uninterrupted treatment or earlier if the disease progresses and the conditions of the managed access agreement for pembrolizumab are followed TA523: Midostaurin for Midostaurin is recommended, within its Red No cost impact to CCGs as NHS England untreated acute myeloid marketing authorisation, as an option in adults commissioned. leukaemia for treating newly diagnosed acute FLT3- mutation-positive myeloid leukaemia with standard daunorubicin and cytarabine as induction therapy, with high-dose cytarabine as consolidation therapy, and alone after complete response as maintenance therapy. -

PIONEER: a Randomized, Double-Blind, Placebo-Controlled, Phase 2 Study of Avapritinib in Patients with Indolent Or Smoldering Sy
PIONEER: A Randomized, Double-Blind, Placebo-Controlled, Phase 2 Study of Avapritinib in Patients with Indolent or Smoldering Systemic Mastocytosis with Symptoms Inadequately Controlled with Standard Therapy Cem Akin, Hanneke Oude Elberink, Jason Gotlib, Vito Sabato, Karin Hartmann, Sigurd Broesby-Olsen, Mariana Castells, Michael W. Deininger, Mark L. Heaney, Tracy I. George, Deepti Radia, Massimo Triggiani, Paul van Daele, Daniel J. DeAngelo, Oleg Schmidt-Kittler, Hui-Min Lin, Andrew Morrison, Brenton Mar, Frank Siebenhaar, Marcus Maurer American Academy of Allergy Asthma and Immunology Annual Meeting March 16, 2020 Disclosures • Investigator: Blueprint Medicines’ ongoing Phase 2 PIONEER trial in indolent and smoldering systemic mastocytosis • Consultant: Blueprint Medicines, Novartis • AYVAKIT™ (avapritinib) is approved by the FDA for the treatment of adults with unresectable or metastatic gastrointestinal stromal tumor (GIST) harboring a platelet-derived growth factor receptor alpha (PDGFRA) exon 18 mutation, including PDGFRA D842V mutations, in the United States. Avapritinib has not been approved by the FDA or any other health authority for use in the United States for any other indication or in any other jurisdiction for any indication. • All data in this presentation are based on a cut-off date of December 27, 2019 unless otherwise specified. 2 Systemic mastocytosis (SM) is a clonal mast cell (MC) neoplasm driven by KIT D816V SM Prevalence of ~1:10,000 ~32,000 estimated in US KIT D816V ~5% Advanced SM Organ damage and decreased survival -

Leukemia Insights Newsletter September 2019 MPN
SEPTEMBER)2019 In this month’s Leukemia Insights newsletter, written by Prithviraj Bose, MD, and Srdan Verstovsek, PhD, MD, and sponsoreD in part by the Charif Souki Cancer Research ABOUT)MyMDAnderson FunD, we summarize the investigational approaches available for patients with myeloproliferative neoplasms (MPN) at MD AnDerson Cancer Center. myMDAnderson is`a`seCure,` personalized`web`site`helping` community`physicians`expedite`patient` referrals,`as`well`as`improve`Continuity` Innovative)Treatment)Strategies)for) of`Care`through`information`acCess` Classic)and)Atypical)Myeloproliferative) and`streamlined`CommuniCations.` PhysiCians`who`have`referred`patients` Neoplasms)(MPN) to`MD`Anderson`or`plan`to`do`so,`Can` utilize`the`HIPAA`Compliant`features In August 2019, the FDA approved the JAK2 inhibitor fedratinib for the of`myMDAnderson`to: treatment of patients with myelofibrosis (MF), only the second drug approval in the classiC myeloproliferative neoplasm (MPN) space after • Refer`a`patient ruxolitinib was approved for MF in 2011 and polyCythemia vera (PV) in • View`your`patient's`appointments` 2014. Hopefully, this is the first of many regulatory approvals as a AcCess`patient`reports number of investigational agents appear promising in reCent and • Send`and`reCeive`seCure` ongoing trials. Below, we summarize the investigational approaches messages available for patients with MPN at MD Anderson. 1.)Pemigatinib for)myeloid/lymphoid)neoplasms)with)FGFR1 JOIN)THE)COVERSATION rearrangement)(NCT03011372) Connect+with+us. Myeloid/lymphoid neoplasms with a rearrangement of the FGFR1 gene loCated at 8p11.2 represent an exCeedingly rare but aggressive malignanCy with no standard treatment options and a dismal prognosis. Eosinophilia is usually present and should trigger testing for rearrangements involving FGFR1, and also PDGFRα and PDGFRβ. -

Samaritan Fund
Items supported by the Samaritan Fund (a) Non-drug Items supported by the Fund (b) Other items supported by the Samaritan Fund Mechanism (c) Self-financed Drugs supported by the Samaritan Fund (SF) and Community Care Fund (CCF) Medical Assistance Programme (First Phase Programme) (for specified self- financed cancer drugs) (a) Non-drug Items supported by the Fund 1. Percutaneous Transluminal Coronary Angioplasty (PTCA) and other consumables for interventional cardiology 2. Cardiac Pacemakers 3. Myoelectric Prosthesis 4. Custom-made Prosthesis 5. Appliances for prosthetic and orthotic services, physiotherapy and occupational therapy services (e.g. prosthesis) 6. Home use equipment and appliances (e.g. wheelchair, replacement of external speech processor for patients done with cochlear implant) 7. Gamma knife surgery 8. Harvesting of marrow in a foreign country for marrow transplant The Fund will only support the model which can meet the basic medical needs of the patients. (b) Other items supported by the Samaritan Fund Mechanism 1. Positron Emission Tomography (PET) service (c) Drugs supported by the Samaritan Fund The following specific self-financed drugs are supported by the Samaritan Fund: Item Drug Types of Clinical indications diseases 1 Abatacept Rheumatology Rheumatoid arthritis 2a Adalimumab Dermatology Severe psoriasis 2b Ophthalmology Non-infectious intermediate, posterior and panuveitis 2c Paediatric chronic non-infectious anterior uveitis 2d Rheumatology Ankylosing spondylitis 2e Juvenile idiopathic arthritis 2f Psoriatic -

GIST): Efficacy and Safety Data from Phase 3 VOYAGER Study 3461883 Yoon-Koo Kang1, Suzanne George2, Robin L
Avapritinib vs Regorafenib in Patients With Locally Advanced Unresectable or Metastatic Gastrointestinal Stromal Tumor Abstract (GIST): Efficacy and Safety Data From Phase 3 VOYAGER Study 3461883 Yoon-Koo Kang1, Suzanne George2, Robin L. Jones3, Piotr Rutkowski4, Lin Shen5, Olivier Mir6, Shreyaskumar Patel7, Yongjian Zhou8, Margaret von Mehren9, Peter Hohenberger10, Victor Villalobos11, Mehdi Brahmi12, William D. Tap13, Jonathan Trent14, Maria A. Pantaleo15, Patrick Schöffski16, Kevin He17, Paggy Hew17, Kate Newberry17, Maria Roche17, Michael Heinrich18, Sebastian Bauer19 1Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea; 2Sarcoma Center, Dana Farber Cancer Institute, Boston, MA, USA; 3Royal Marsden Hospital and Institute of Cancer Research, London, UK; 4Maria Sklodowska-Curie National Research Institute of Oncology, Warsaw, Poland; 5Peking University Cancer Hospital and Institute, Beijing, China; 6Institut Gustave Roussy, Villejuif, France; 7MD Anderson Cancer Center, Houston, TX, USA; 8Fujian Medical University Union Hospital, Fuzhou, China; 9Fox Chase Cancer Center, Philadelphia, PA, USA; 10Medical Faculty Mannheim (UMM), University of Heidelberg, Mannheim, Germany; 11University of Colorado Denver – Anschutz Medical Campus, Aurora, CO, USA; 12Centre Léon Bérard, Lyon, France; 13Memorial Sloan Kettering Cancer Center and Weill Cornell Medical College, New York, NY, USA; 14University of Miami-Sylvester Comprehensive Cancer Center, Miami, FL, USA; 15University of Bologna, Bologna, Italy; 16University Hospitals