(12) Patent Application Publication (10) Pub. No.: US 2013/0274315 A1 Birrer Et Al
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Binnenwerk Cindy Postma.Indd
CHAPTER 6 Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression Gut 2009, 58: 79-89 Beatriz Carvalho Cindy Postma Sandra Mongera Erik Hopmans Sharon Diskin Mark A. van de Wiel Wim van Criekinge Olivier Thas Anja Matthäi Miguel A. Cuesta Jochim S. Terhaar sive Droste Mike Craanen Evelin Schröck Bauke Ylstra Gerrit A. Meijer 104 | Chapter 6 Abstract Objective: This study aimed to identify the oncogenes at 20q involved in colorectal adenoma to carcinoma progression by measuring the effect of 20q gain on mRNA expression of genes in this amplicon. Methods: Segmentation of DNA copy number changes on 20q was performed by array CGH in 34 non-progressed colorectal adenomas, 41 progressed adenomas (i.e. adenomas that present a focus of cancer) and 33 adenocarcinomas. Moreover, a robust analysis of altered expression of genes in these segments was performed by microarray analysis in 37 adenomas and 31 adenocarcinomas. Protein expression was evaluated by immunohistochemistry on tissue microarrays. Results: The genes C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCFL5, mapping at 20q were signifi cantly overexpressed in carcinomas compared to adenomas as consequence of copy number gain of 20q. Conclusion: This approach revealed C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCFL5 genes to be important in chromosomal instability-related adenoma to carcinoma progression. These genes therefore may serve as highly specifi c biomarkers for colorectal cancer with potential clinical applications. Putative oncogenes at chromosome 20q in colorectal carcinogenesis | 105 Introduction The majority of cancers are epithelial in origin and arise through a stepwise progression from normal cells, through dysplasia, into malignant cells that invade surrounding tissues and have metastatic potential. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signifcance and Impact on Clinical Features
Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signicance and Impact on Clinical Features Fadwa Said Abdelazim Mohamed ( [email protected] ) Cairo University Roxan E Shak National Cancer Institute, Cairo University Naglaa M. Hassan National Cancer Institute, Cairo University Research article Keywords: Acute myeloid leukemia, RUNX1, mutation, gene expression, Egypt Posted Date: April 7th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-20926/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/20 Abstract Background Acute myeloid leukemia represents the highest percentage of all adult acute leukemia variants. Runt-related transcription factor1 (RUNX1), a transcription factor with a known tumor suppressor function was recently reported as a tumor promotor in AML. We investigated the role of RUNX1 geneexpression levelin Egyptian AML patients and delineateits clinical signicance. Methods This study recruited 91 AML patients that were recently diagnosed at our hospital with 14 healthy age- and sex-matched donors of bone marrow transplantation unit. We measured RUNX1 gene expression level using reverse transcription–quantitative polymerase chain reaction. Results We found that RUNX1 gene expression level was signicantly higher compared to the control group (p < 0.001). Patients with FLT3 mutations had the higher expression level of RUNX1 (p=0.023). The male patients expressed signicantly higher level of RUNX1 (p=0.046). Conclusion The RUNX1 gene expression may serve as a diagnostic marker in Egyptian AML patients. Its relation to FLT3 may give clue that patients carrying this mutation may benet fromnew treatments that target RUNX1 in the future. -

Table SI. Genes Upregulated ≥ 2-Fold by MIH 2.4Bl Treatment Affymetrix ID
Table SI. Genes upregulated 2-fold by MIH 2.4Bl treatment Fold UniGene ID Description Affymetrix ID Entrez Gene Change 1558048_x_at 28.84 Hs.551290 231597_x_at 17.02 Hs.720692 238825_at 10.19 93953 Hs.135167 acidic repeat containing (ACRC) 203821_at 9.82 1839 Hs.799 heparin binding EGF like growth factor (HBEGF) 1559509_at 9.41 Hs.656636 202957_at 9.06 3059 Hs.14601 hematopoietic cell-specific Lyn substrate 1 (HCLS1) 202388_at 8.11 5997 Hs.78944 regulator of G-protein signaling 2 (RGS2) 213649_at 7.9 6432 Hs.309090 serine and arginine rich splicing factor 7 (SRSF7) 228262_at 7.83 256714 Hs.127951 MAP7 domain containing 2 (MAP7D2) 38037_at 7.75 1839 Hs.799 heparin binding EGF like growth factor (HBEGF) 224549_x_at 7.6 202672_s_at 7.53 467 Hs.460 activating transcription factor 3 (ATF3) 243581_at 6.94 Hs.659284 239203_at 6.9 286006 Hs.396189 leucine rich single-pass membrane protein 1 (LSMEM1) 210800_at 6.7 1678 translocase of inner mitochondrial membrane 8 homolog A (yeast) (TIMM8A) 238956_at 6.48 1943 Hs.741510 ephrin A2 (EFNA2) 242918_at 6.22 4678 Hs.319334 nuclear autoantigenic sperm protein (NASP) 224254_x_at 6.06 243509_at 6 236832_at 5.89 221442 Hs.374076 adenylate cyclase 10, soluble pseudogene 1 (ADCY10P1) 234562_x_at 5.89 Hs.675414 214093_s_at 5.88 8880 Hs.567380; far upstream element binding protein 1 (FUBP1) Hs.707742 223774_at 5.59 677825 Hs.632377 small nucleolar RNA, H/ACA box 44 (SNORA44) 234723_x_at 5.48 Hs.677287 226419_s_at 5.41 6426 Hs.710026; serine and arginine rich splicing factor 1 (SRSF1) Hs.744140 228967_at 5.37 -

Supplemental Information
Supplemental information Dissection of the genomic structure of the miR-183/96/182 gene. Previously, we showed that the miR-183/96/182 cluster is an intergenic miRNA cluster, located in a ~60-kb interval between the genes encoding nuclear respiratory factor-1 (Nrf1) and ubiquitin-conjugating enzyme E2H (Ube2h) on mouse chr6qA3.3 (1). To start to uncover the genomic structure of the miR- 183/96/182 gene, we first studied genomic features around miR-183/96/182 in the UCSC genome browser (http://genome.UCSC.edu/), and identified two CpG islands 3.4-6.5 kb 5’ of pre-miR-183, the most 5’ miRNA of the cluster (Fig. 1A; Fig. S1 and Seq. S1). A cDNA clone, AK044220, located at 3.2-4.6 kb 5’ to pre-miR-183, encompasses the second CpG island (Fig. 1A; Fig. S1). We hypothesized that this cDNA clone was derived from 5’ exon(s) of the primary transcript of the miR-183/96/182 gene, as CpG islands are often associated with promoters (2). Supporting this hypothesis, multiple expressed sequences detected by gene-trap clones, including clone D016D06 (3, 4), were co-localized with the cDNA clone AK044220 (Fig. 1A; Fig. S1). Clone D016D06, deposited by the German GeneTrap Consortium (GGTC) (http://tikus.gsf.de) (3, 4), was derived from insertion of a retroviral construct, rFlpROSAβgeo in 129S2 ES cells (Fig. 1A and C). The rFlpROSAβgeo construct carries a promoterless reporter gene, the β−geo cassette - an in-frame fusion of the β-galactosidase and neomycin resistance (Neor) gene (5), with a splicing acceptor (SA) immediately upstream, and a polyA signal downstream of the β−geo cassette (Fig. -

Identification of Transcriptomic Differences Between Lower
International Journal of Molecular Sciences Article Identification of Transcriptomic Differences between Lower Extremities Arterial Disease, Abdominal Aortic Aneurysm and Chronic Venous Disease in Peripheral Blood Mononuclear Cells Specimens Daniel P. Zalewski 1,*,† , Karol P. Ruszel 2,†, Andrzej St˛epniewski 3, Dariusz Gałkowski 4, Jacek Bogucki 5 , Przemysław Kołodziej 6 , Jolanta Szyma ´nska 7 , Bartosz J. Płachno 8 , Tomasz Zubilewicz 9 , Marcin Feldo 9,‡ , Janusz Kocki 2,‡ and Anna Bogucka-Kocka 1,‡ 1 Chair and Department of Biology and Genetics, Medical University of Lublin, 4a Chod´zkiSt., 20-093 Lublin, Poland; [email protected] 2 Chair of Medical Genetics, Department of Clinical Genetics, Medical University of Lublin, 11 Radziwiłłowska St., 20-080 Lublin, Poland; [email protected] (K.P.R.); [email protected] (J.K.) 3 Ecotech Complex Analytical and Programme Centre for Advanced Environmentally Friendly Technologies, University of Marie Curie-Skłodowska, 39 Gł˛ebokaSt., 20-612 Lublin, Poland; [email protected] 4 Department of Pathology and Laboratory Medicine, Rutgers-Robert Wood Johnson Medical School, One Robert Wood Johnson Place, New Brunswick, NJ 08903-0019, USA; [email protected] 5 Chair and Department of Organic Chemistry, Medical University of Lublin, 4a Chod´zkiSt., Citation: Zalewski, D.P.; Ruszel, K.P.; 20-093 Lublin, Poland; [email protected] St˛epniewski,A.; Gałkowski, D.; 6 Laboratory of Diagnostic Parasitology, Chair and Department of Biology and Genetics, Medical University of Bogucki, J.; Kołodziej, P.; Szyma´nska, Lublin, 4a Chod´zkiSt., 20-093 Lublin, Poland; [email protected] J.; Płachno, B.J.; Zubilewicz, T.; Feldo, 7 Department of Integrated Paediatric Dentistry, Chair of Integrated Dentistry, Medical University of Lublin, M.; et al. -

Antigen-Specific Memory CD4 T Cells Coordinated Changes in DNA
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology The Journal of Immunology published online 18 March 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/03/17/jimmun ol.1202267 Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto, Katsumi Ogoshi, Atsushi Sasaki, Jun Abe, Wei Qu, Yoichiro Nakatani, Budrul Ahsan, Kenshiro Oshima, Francis H. W. Shand, Akio Ametani, Yutaka Suzuki, Shuichi Kaneko, Takashi Wada, Masahira Hattori, Sumio Sugano, Shinichi Morishita and Kouji Matsushima J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Author Choice option Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Freely available online through http://www.jimmunol.org/content/suppl/2013/03/18/jimmunol.120226 7.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Author Choice Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. Published March 18, 2013, doi:10.4049/jimmunol.1202267 The Journal of Immunology Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto,*,†,‡ Katsumi Ogoshi,* Atsushi Sasaki,† Jun Abe,* Wei Qu,† Yoichiro Nakatani,† Budrul Ahsan,x Kenshiro Oshima,† Francis H. -

ITRAQ-Based Quantitative Proteomic Analysis of Processed Euphorbia Lathyris L
Zhang et al. Proteome Science (2018) 16:8 https://doi.org/10.1186/s12953-018-0136-6 RESEARCH Open Access ITRAQ-based quantitative proteomic analysis of processed Euphorbia lathyris L. for reducing the intestinal toxicity Yu Zhang1, Yingzi Wang1*, Shaojing Li2*, Xiuting Zhang1, Wenhua Li1, Shengxiu Luo1, Zhenyang Sun1 and Ruijie Nie1 Abstract Background: Euphorbia lathyris L., a Traditional Chinese medicine (TCM), is commonly used for the treatment of hydropsy, ascites, constipation, amenorrhea, and scabies. Semen Euphorbiae Pulveratum, which is another type of Euphorbia lathyris that is commonly used in TCM practice and is obtained by removing the oil from the seed that is called paozhi, has been known to ease diarrhea. Whereas, the mechanisms of reducing intestinal toxicity have not been clearly investigated yet. Methods: In this study, the isobaric tags for relative and absolute quantitation (iTRAQ) in combination with the liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomic method was applied to investigate the effects of Euphorbia lathyris L. on the protein expression involved in intestinal metabolism, in order to illustrate the potential attenuated mechanism of Euphorbia lathyris L. processing. Differentially expressed proteins (DEPs) in the intestine after treated with Semen Euphorbiae (SE), Semen Euphorbiae Pulveratum (SEP) and Euphorbiae Factor 1 (EFL1) were identified. The bioinformatics analysis including GO analysis, pathway analysis, and network analysis were done to analyze the key metabolic pathways underlying the attenuation mechanism through protein network in diarrhea. Western blot were performed to validate selected protein and the related pathways. Results: A number of differentially expressed proteins that may be associated with intestinal inflammation were identified. -

Dissertation
Dissertation submitted to the Combined Faculty of Natural Sciences and Mathematics of the Ruperto Carola University Heidelberg, Germany for the degree of Doctor of Natural Sciences Presented by: Arlou Kristina J. Angeles, M.Sc. born in Manila, Philippines Oral examination: 19 July 2019 The ERG-driven long non-coding RNA LINC00920 promotes cell proliferation and migration in prostate cancer cells by modulating FOXO activity through a direct interaction with 14-3-3ε Referees: PD Dr. Odilia Popanda Prof. Dr. Holger Sültmann Declaration I hereby declare that the submitted dissertation entitled “The ERG-driven long non-coding RNA LINC00920 promotes cell proliferation and migration in prostate cancer cells by modulating FOXO activity through a direct interaction with 14-3-3ε” is the product of my own work and that all assistance received in preparing this thesis and manuscript have been acknowledged. I have not applied to be examined at any other institution, nor have I submitted this dissertation to any other faculty. Furthermore, I took reasonable care to ensure that the work is original, and, to the best of my knowledge, does not breach copyright law, and has not been taken from other sources except where such work has been cited and acknowledged within the text. ________________________ ________________________ Place, Date Arlou Kristina Angeles Summary This thesis describes LINC00920, a tumor-associated lncRNA identified in the transcriptome dataset of the International Cancer Genome Consortium-Early Onset Prostate Cancer (ICGC-EOPC) cohort. SiRNA-mediated knockdown of LINC00920 negatively affected proliferation, colony formation, and migration of PC-3 prostate cancer cells. Gene set enrichment analysis of microarray expression data revealed perturbation of pathways related to cell cycle, cell division, apoptosis, and cell movement. -

Virulent Diuraphis Noxia Aphids Over-Express Calcium Signaling Proteins to Overcome Defenses of Aphid-Resistant Wheat Plants
RESEARCH ARTICLE Virulent Diuraphis noxia Aphids Over-Express Calcium Signaling Proteins to Overcome Defenses of Aphid-Resistant Wheat Plants Deepak K. Sinha1,2, Predeesh Chandran2, Alicia E. Timm2, Lina Aguirre-Rojas2,C. Michael Smith2* 1 International Centre for Genetic Engineering and Biotechnology, New Delhi 110067, India, 2 Department of Entomology, Kansas State University, Manhattan, Kansas 66506–4004, United States of America * [email protected] Abstract The Russian wheat aphid, Diuraphis noxia, an invasive phytotoxic pest of wheat, Triticum aestivum, and barley, Hordeum vulgare, causes huge economic losses in Africa, South America, and North America. Most acceptable and ecologically beneficial aphid manage- OPEN ACCESS ment strategies include selection and breeding of D. noxia-resistant varieties, and numer- Citation: Sinha DK, Chandran P, Timm AE, Aguirre- ous D. noxia resistance genes have been identified in T. aestivum and H. vulgare. North Rojas L, Smith CM (2016) Virulent Diuraphis noxia American D. noxia biotype 1 is avirulent to T. aestivum varieties possessing Dn4 or Dn7 Aphids Over-Express Calcium Signaling Proteins to genes, while biotype 2 is virulent to Dn4 and avirulent to Dn7. The current investigation uti- Overcome Defenses of Aphid-Resistant Wheat Plants. PLoS ONE 11(1): e0146809. doi:10.1371/ lized next-generation RNAseq technology to reveal that biotype 2 over expresses proteins journal.pone.0146809 involved in calcium signaling, which activates phosphoinositide (PI) metabolism. Calcium Editor: Guangxiao Yang, Huazhong University of signaling proteins comprised 36% of all transcripts identified in the two D. noxia biotypes. Science & Technology(HUST), CHINA Depending on plant resistance gene-aphid biotype interaction, additional transcript groups Received: September 3, 2015 included those involved in tissue growth; defense and stress response; zinc ion and related cofactor binding; and apoptosis. -
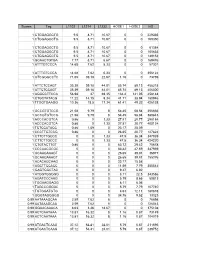
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

Looking for Missing Proteins in the Proteome Of
Looking for Missing Proteins in the Proteome of Human Spermatozoa: An Update Yves Vandenbrouck, Lydie Lane, Christine Carapito, Paula Duek, Karine Rondel, Christophe Bruley, Charlotte Macron, Anne Gonzalez de Peredo, Yohann Coute, Karima Chaoui, et al. To cite this version: Yves Vandenbrouck, Lydie Lane, Christine Carapito, Paula Duek, Karine Rondel, et al.. Looking for Missing Proteins in the Proteome of Human Spermatozoa: An Update. Journal of Proteome Research, American Chemical Society, 2016, 15 (11), pp.3998-4019. 10.1021/acs.jproteome.6b00400. hal-02191502 HAL Id: hal-02191502 https://hal.archives-ouvertes.fr/hal-02191502 Submitted on 19 Mar 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Journal of Proteome Research 1 2 3 Looking for missing proteins in the proteome of human spermatozoa: an 4 update 5 6 Yves Vandenbrouck1,2,3,#,§, Lydie Lane4,5,#, Christine Carapito6, Paula Duek5, Karine Rondel7, 7 Christophe Bruley1,2,3, Charlotte Macron6, Anne Gonzalez de Peredo8, Yohann Couté1,2,3, 8 Karima Chaoui8, Emmanuelle Com7, Alain Gateau5, AnneMarie Hesse1,2,3, Marlene 9 Marcellin8, Loren Méar7, Emmanuelle MoutonBarbosa8, Thibault Robin9, Odile Burlet- 10 Schiltz8, Sarah Cianferani6, Myriam Ferro1,2,3, Thomas Fréour10,11, Cecilia Lindskog12,Jérôme 11 1,2,3 7,§ 12 Garin , Charles Pineau .