Nvpou4/Brain3 Functions As a Terminal Selector Gene in the Nervous System of The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sexual Reproduction 7.2: Meiosis 7.3: Errors in Meiosis
Concepts of Biology Chapter 6 | Reproduction at the Cellular Level 135 6 | REPRODUCTION AT THE CELLULAR LEVEL Figure 6.1 A sea urchin begins life as a single cell that (a) divides to form two cells, visible by scanning electron microscopy. After four rounds of cell division, (b) there are 16 cells, as seen in this SEM image. After many rounds of cell division, the individual develops into a complex, multicellular organism, as seen in this (c) mature sea urchin. (credit a: modification of work by Evelyn Spiegel, Louisa Howard; credit b: modification of work by Evelyn Spiegel, Louisa Howard; credit c: modification of work by Marco Busdraghi; scale-bar data from Matt Russell) Chapter Outline 6.1: The Genome 6.2: The Cell Cycle 6.3: Cancer and the Cell Cycle 6.4: Prokaryotic Cell Division Introduction The individual sexually reproducing organism—including humans—begins life as a fertilized egg, or zygote. Trillions of cell divisions subsequently occur in a controlled manner to produce a complex, multicellular human. In other words, that original single cell was the ancestor of every other cell in the body. Once a human individual is fully grown, cell reproduction is still necessary to repair or regenerate tissues. For example, new blood and skin cells are constantly being produced. All multicellular organisms use cell division for growth, and in most cases, the maintenance and repair of cells and tissues. Single-celled organisms use cell division as their method of reproduction. 6.1 | The Genome By the end of this section, you will be able to: • Describe the prokaryotic and eukaryotic genome • Distinguish between chromosomes, genes, and traits The continuity of life from one cell to another has its foundation in the reproduction of cells by way of the cell cycle. -

The Evolution of Siphonophore Tentilla for Specialized Prey Capture in the Open Ocean
The evolution of siphonophore tentilla for specialized prey capture in the open ocean Alejandro Damian-Serranoa,1, Steven H. D. Haddockb,c, and Casey W. Dunna aDepartment of Ecology and Evolutionary Biology, Yale University, New Haven, CT 06520; bResearch Division, Monterey Bay Aquarium Research Institute, Moss Landing, CA 95039; and cEcology and Evolutionary Biology, University of California, Santa Cruz, CA 95064 Edited by Jeremy B. C. Jackson, American Museum of Natural History, New York, NY, and approved December 11, 2020 (received for review April 7, 2020) Predator specialization has often been considered an evolutionary makes them an ideal system to study the relationships between “dead end” due to the constraints associated with the evolution of functional traits and prey specialization. Like a head of coral, a si- morphological and functional optimizations throughout the organ- phonophore is a colony bearing many feeding polyps (Fig. 1). Each ism. However, in some predators, these changes are localized in sep- feeding polyp has a single tentacle, which branches into a series of arate structures dedicated to prey capture. One of the most extreme tentilla. Like other cnidarians, siphonophores capture prey with cases of this modularity can be observed in siphonophores, a clade of nematocysts, harpoon-like stinging capsules borne within special- pelagic colonial cnidarians that use tentilla (tentacle side branches ized cells known as cnidocytes. Unlike the prey-capture apparatus of armed with nematocysts) exclusively for prey capture. Here we study most other cnidarians, siphonophore tentacles carry their cnidocytes how siphonophore specialists and generalists evolve, and what mor- in extremely complex and organized batteries (3), which are located phological changes are associated with these transitions. -

Feeding-Dependent Tentacle Development in the Sea Anemone Nematostella Vectensis ✉ Aissam Ikmi 1,2 , Petrus J
ARTICLE https://doi.org/10.1038/s41467-020-18133-0 OPEN Feeding-dependent tentacle development in the sea anemone Nematostella vectensis ✉ Aissam Ikmi 1,2 , Petrus J. Steenbergen1, Marie Anzo 1, Mason R. McMullen2,3, Anniek Stokkermans1, Lacey R. Ellington2 & Matthew C. Gibson2,4 In cnidarians, axial patterning is not restricted to embryogenesis but continues throughout a prolonged life history filled with unpredictable environmental changes. How this develop- 1234567890():,; mental capacity copes with fluctuations of food availability and whether it recapitulates embryonic mechanisms remain poorly understood. Here we utilize the tentacles of the sea anemone Nematostella vectensis as an experimental paradigm for developmental patterning across distinct life history stages. By analyzing over 1000 growing polyps, we find that tentacle progression is stereotyped and occurs in a feeding-dependent manner. Using a combination of genetic, cellular and molecular approaches, we demonstrate that the crosstalk between Target of Rapamycin (TOR) and Fibroblast growth factor receptor b (Fgfrb) signaling in ring muscles defines tentacle primordia in fed polyps. Interestingly, Fgfrb-dependent polarized growth is observed in polyp but not embryonic tentacle primordia. These findings show an unexpected plasticity of tentacle development, and link post-embryonic body patterning with food availability. 1 Developmental Biology Unit, European Molecular Biology Laboratory, 69117 Heidelberg, Germany. 2 Stowers Institute for Medical Research, Kansas City, MO 64110, -

Hazardous Marine Life
JOIN US AT DAN.ORG HAZARDOUS MARINE LIFE STINGS Cnidarians (nematocyst-carrying species) are HEALTH & DIVING REFERENCE SERIES responsible for more evenomations than any other marine phylum. These organisms contain stinging cells called cnidocyte that excel at venom delivery. 6 West Colony Place Durham, NC 27705 USA HAZARDOUS JELLYFISH PHONE: +1-919-684-2948 Of all the cnidarians, jellyfish cause the most MARINE LIFE: DAN EMERGENCY HOTLINE: +1-919-684-9111 frequent and severe human injuries. These result LEARN MORE AT DAN.ORG/HEALTH STINGS from direct contact with tentacles, and though painful, are not typically life threatening. PORTUGUESE MAN-OF-WAR Portuguese man-of-wars are free-floating cnidarians characterized by blue gas-filled bladders and long tentacles that drift on the ocean’s surface. Contact with a man-of-war’s Part #: 013-1040 Rev. 7.27.15 tentacles can cause significant pain and systemic symptoms. Their tentacles contain cnidocytes that deliver a potent proteic neurotoxin. Despite its resemblance to jellyfish, Portuguese man-of-war are more closely related to fire coral than to true jellyfish. HYDROIDS Although hydroids look like plants, these feathery cnidarians are actually a colony of small zooids that work together as a functioning animal. Like all cnidarians, these animals are armed with stinging cnidocytes. Hydroids, Portuguese man-of-war and fire coral belong to the same family, and contact with them can cause very similar reactions. Granted, their smaller contact area usually results in more localized and less dramatic reactions. FIRE CORAL Fire coral are colonial marine cnidarians that cause a mild to moderate burning reaction when touched. -
Cnidarians Are Some of the Most Conspicuous and Colorful Tidepool Animals, Yet They Are Often Overlooked and Unappreciated
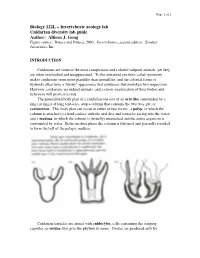
Page 1 of 1 Biology 122L – Invertebrate zoology lab Cnidarian diversity lab guide Author: Allison J. Gong Figure source: Brusca and Brusca, 2003. Invertebrates, second edition. Sinauer Associates, Inc. INTRODUCTION Cnidarians are some of the most conspicuous and colorful tidepool animals, yet they are often overlooked and unappreciated. To the untrained eye their radial symmetry makes cnidarians seem more plantlike than animallike, and the colonial forms of hydroids often have a "bushy" appearance that reinforces that mistaken first impression. However, cnidarians are indeed animals, and a closer examination of their bodies and behaviors will prove it to you. The generalized body plan of a cnidarian consists of an oral disc surrounded by a ring (or rings) of long tentacles, atop a column that contains the two-way gut, or coelenteron. This body plan can occur in either of two forms: a polyp, in which the column is attached to a hard surface with the oral disc and tentacles facing into the water; and a medusa, in which the column is (usually) unattached and the entire organism is surrounded by water. In the medusa phase the column is flattened and generally rounded to form the bell of the pelagic medusa. Cnidarian tentacles are armed with cnidocytes, cells containing the stinging capsules, or cnidae, that give the phylum its name. Cnidae are produced only by Page 2 of 2 cnidarians, although they can occasionally be found in the cerata of nudibranch molluscs that feed on cnidarians: through a process that is not understood, some nudibranchs are able to ingest cnidae from their prey and sequester them, unfired, in their cerata for defense against their own predators. -

How Cnidaria Got Its Cnidocysts
tems: ys Op l S e a n A ic c g c o l e s o i s Shostak, Biol syst Open Access 2015, 4:2 B Biological Systems: Open Access DOI: 10.4172/2329-6577.1000139 ISSN: 2329-6577 Review Article Open Access How Cnidaria Got Its Cnidocysts Stanley Shostak* Department of Biological Sciences, University of Pittsburgh, USA *Corresponding author: Stanley Shostak, Department of Biological Sciences, University of Pittsburgh, USA, Tel: 0114129156595, E-mail: [email protected] Received date: May 06, 2015; Accepted date: Aug 03, 2015; Published date: Aug 11, 2015 Copyright: © 2015 Shostak S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract A complex hypothesis is offered for the origins of cnidarian cnidocysts through symbiogeny. The two-part hypothetical pathway links the origins of tissues through an early amalgamation of amoebic and epithelial cells to and the later introduction of an extrusion apparatus from bacterial parasites. The first part of the hypothesis is based on evidence for morphological, molecular, and developmental similarities of cnidarians and myxozoans indicative of common ancestry. Support is drawn from Ediacaran fossils suggesting that stem-metazoans consisted of symbiogenic pairs of epithelial-like shells enclosing amoeba-like cells. The amoeba-like cells would have evolved into germ cells and cells differentiating as or inducing nerve, muscle, and gland cells. The second part of the hypothesis proposes that cnidocysts evolved in a cnidarian/myxozoan branch of the metazoan tree through the horizontal transfer of bacterial genes encoding an extrusion apparatus to proto-cnidarian amoebic cells and consequently to the Cnidarian germ line. -

Six Major Steps in Animal Evolution: Are We Derived Sponge Larvae?
EVOLUTION & DEVELOPMENT 10:2, 241–257 (2008) Six major steps in animal evolution: are we derived sponge larvae? Claus Nielsen Zoological Museum (The Natural History Museum of Denmark, University of Copenhagen), Universitetsparken 15, DK-2100 Copenhagen, Denmark Correspondence (email: [email protected]) SUMMARY A review of the old and new literature on animal became sexually mature, and the adult sponge-stage was morphology/embryology and molecular studies has led me to abandoned in an extreme progenesis. This eumetazoan the following scenario for the early evolution of the metazoans. ancestor, ‘‘gastraea,’’ corresponds to Haeckel’s gastraea. The metazoan ancestor, ‘‘choanoblastaea,’’ was a pelagic Trichoplax represents this stage, but with the blastopore spread sphere consisting of choanocytes. The evolution of multicellularity out so that the endoderm has become the underside of the enabled division of labor between cells, and an ‘‘advanced creeping animal. Another lineage developed a nervous system; choanoblastaea’’ consisted of choanocytes and nonfeeding cells. this ‘‘neurogastraea’’ is the ancestor of the Neuralia. Cnidarians Polarity became established, and an adult, sessile stage have retained this organization, whereas the Triploblastica developed. Choanocytes of the upper side became arranged in (Ctenophora1Bilateria), have developed the mesoderm. The a groove with the cilia pumping water along the groove. Cells bilaterians developed bilaterality in a primitive form in the overarched the groove so that a choanocyte chamber was Acoelomorpha and in an advanced form with tubular gut and formed, establishing the body plan of an adult sponge; the pelagic long Hox cluster in the Eubilateria (Protostomia1Deuterostomia). larval stage was retained but became lecithotrophic. The It is indicated that the major evolutionary steps are the result of sponges radiated into monophyletic Silicea, Calcarea, and suites of existing genes becoming co-opted into new networks Homoscleromorpha. -

Phylum Cnidaria
Name___________________________________ Biology II --- May 2012 Phylum Cnidaria Cnidarians include corals, jellyfish, sea anemones, and hydra. Jellyfish are fish-eating animals that float in the sea - only a few jellyfish live in fresh water. They have soft bodies and long, stinging, poisonous tentacles that they use to catch fish. Label and color the tentacles green. The tentacles hang down from an umbrella-shaped medusa. Label the medusa. The jellyfish has two cell layers --- an outer ectoderm and an inner endoderm. Label and color the ectoderm red and the endoderm orange. Between these layers is a jellylike material called mesoglea. Label and color the mesoglea pink. The mouth is located on the underside in the center of the tentacles. Label the mouth. The mouth opens into the gastrovascular cavity where their prey is digested. Label and color the gastrovascular cavity tan. Venom or a paralyzing poison is sent out through stinging cells called cnidocytes. Label and color the cnidocyte yellow. Inside the cnidocytes, is a coiled, poisonous thread that can shoot out to paralyze prey with poison. This harpoon-like structure is called the nematocyst. Cnidocytes have a trigger that when touched shoot out the nematocyst. Label and color the nematocyst orange. Label the trigger on the cnidocyte. Jellyfish go through two stages in their life cycle --- polyp and medusa. The adult jellyfish has tentacles hanging downward and is called the medusa stage. Label and color the adult medusa stage red. Eggs and sperm are released into the water and join to form a fertilized egg or zygote. Color and label the zygote yellow. -

NF-ᅫᄎB Is Required for Cnidocyte Development in the Sea Anemone
Developmental Biology 373 (2013) 205–215 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/locate/developmentalbiology Evolution of Developmental Control Mechanisms NF-kB is required for cnidocyte development in the sea anemone Nematostella vectensis Francis S. Wolenski a, Cynthia A. Bradham a,b, John R. Finnerty a,b, Thomas D. Gilmore a,n a Boston University, Department of Biology, 5 Cummington Mall, Boston, MA 02215, United States b Program in Bioinformatics, Boston University, Boston, MA 02215, United States article info abstract Article history: The sea anemone Nematostella vectensis (Nv) is a leading model organism for the phylum Cnidaria, Received 17 May 2012 which includes anemones, corals, jellyfishes and hydras. A defining trait across this phylum is the Received in revised form cnidocyte, an ectodermal cell type with a variety of functions including defense, prey capture and 12 September 2012 environmental sensing. Herein, we show that the Nv-NF-kB transcription factor and its inhibitor Accepted 4 October 2012 Nv-IkB are expressed in a subset of cnidocytes in the body column of juvenile and adult anemones. The Available online 12 October 2012 size and distribution of the Nv-NF-kB-positive cnidocytes suggest that they are in a subtype known as Keywords: basitrichous haplonema cnidocytes. Nv-NF-kB is primarily cytoplasmic in cnidocytes in juvenile and NF-kappaB adult animals, but is nuclear when first detected in the 30-h post-fertilization embryo. Morpholino- IkappaB mediated knockdown of Nv-NF-kB expression results in greatly reduced cnidocyte formation in the Nematostella vectensis 5 day-old animal. -

BI 101: Invertebrate Animals
12/4/2013 BI 101: Invertebrate Animals Announcements • Lab tomorrow: Invertebrates ( lab worksheet provided) – 8 am start – No prelab • Holiday: No class Friday! 1 12/4/2013 Classification The three-domains Bacteria Archaea Eukarya The six-kingdom system Bacteria Archaea Protista Plantae Fungi Animalia The traditional five-kingdom system Monera Protista Plantae Fungi Animalia (Forams and Radiolarians) Rhizarians Alveolates Rhodophyta Stramenopile CHLOROPHYTA Euglenozoa AMOEBOZOANS 2 12/4/2013 What are some characteristics animals share? List as many as you can think of. Discuss this in your groups Animal Cell Fungus Cell 3 12/4/2013 Evidence indicates that animals evolved from choanoflagellates (protists) ~ 570 mya • Single cells • Often clonal • Heterotroph • No specialization or coodination between cells Animal Classification 1. DNA sequencing 2. Body Symmetry 3. Presence or absence of body cavity 4. Embyonic Development 4 12/4/2013 Symmetry Body Cavity Most bilateral animals have body cavities – Body cavities are fluid-filled cavities between the digestive tube and the outer body wall – Functions: • skeleton, providing support for the body and a framework against which muscles can act • protective buffer between the internal organs and the outside world • They can allow organs to move independently of the body wall 5 12/4/2013 Body Cavity? epidermis gut cavity organs packed between A No coelom gut and body wall (acoelomate animal) Fig. 25-4a, p. 405 Body Cavity? epidermis gut cavity B Pseudocoel (pseudocoelomate animal) unlined body cavity around gut Fig. 25-4b, p. 405 6 12/4/2013 Body Cavity? gut epidermis cavity C Coelom body cavity with a lining (dark (coelomate animal) blue) derived from mesoderm Fig. -

Cnidocyte Discharge Is Regulated by Light and Opsin-Mediated Phototransduction
UC Davis UC Davis Previously Published Works Title Cnidocyte discharge is regulated by light and opsin-mediated phototransduction Permalink https://escholarship.org/uc/item/4cm5z4qm Journal BMC Biology, 10(1) ISSN 1741-7007 Authors Plachetzki, David C Fong, Caitlin R Oakley, Todd H Publication Date 2012-03-05 DOI http://dx.doi.org/10.1186/1741-7007-10-17 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Cnidocyte discharge is regulated by light and opsin-mediated phototransduction Plachetzki et al. Plachetzki et al. BMC Biology 2012, 10:17 http://www.biomedcentral.com/1741-7007/10/17 (5 March 2012) Plachetzki et al. BMC Biology 2012, 10:17 http://www.biomedcentral.com/1741-7007/10/17 RESEARCHARTICLE Open Access Cnidocyte discharge is regulated by light and opsin-mediated phototransduction David C Plachetzki1*, Caitlin R Fong2 and Todd H Oakley2 Abstract Background: Cnidocytes, the eponymous cell type of the Cnidaria, facilitate both sensory and secretory functions and are among the most complex animal cell types known. In addition to their structural complexity, cnidocytes display complex sensory attributes, integrating both chemical and mechanical cues from the environment into their discharge behavior. Despite more than a century of work aimed at understanding the sensory biology of cnidocytes, the specific sensory receptor genes that regulate their function remain unknown. Results: Here we report that light also regulates cnidocyte function. We show that non-cnidocyte neurons located in battery complexes of the freshwater polyp Hydra magnipapillata specifically express opsin, cyclic nucleotide gated (CNG) ion channel and arrestin, which are all known components of bilaterian phototransduction cascades. -

CNIDARIA • TISSUE Level of Body Organization
PHYLUM CNIDARIA • TISSUE level of body organization • Middle layer = MESOGLEA = Acellular matrix (Just jelly!) • Diagnostic cell type = CNIDOCYTE It contains the Nematocyst organelle Cnidocyte vs. Nematocyst B C A A = ? ? B = ? C = ? Insert: A Cnidocyte (C) – cell containing a Nematocyst - organelle not yet triggered. E E G M G Cnidarians are DIPLOBLASTIC (2 tissue layers) C = Epidermis (E) & A = Gastrodermis (G) with B = Mesoglea in between the two Phylum Cnidaria Close-up of Nematocysts Specialized cells called cnidocytes contain nematocysts. These are used for anchorage, defense and capture of prey. Cnidarian Life Cycles •Hydrozoa Polyp dominant Medusa does exist (Hydra is cute but odd!) Remember the fire coral! • Scyphozoa Medusa dominant Polyp does exist • Anthozoa Polyp only Do you know the difference between a bud and a gonad? PHYLUM Cnidaria cLASS Hydrozoa Cnidocyte-bearing tentacles, mouth, GVC & bud (branch = asexual reproduction) [fig 2.2] PHYLUM Cnidaria CLASS Hydrozoa Polyp with gonads for sexual reproduction & close-up view of the gonads [fig 2.2] (bumps) Which structure is used for what? PHYLUM Cnidaria CLASS Hydrozoa Obelia colony slide with close-up of the some of the polyps or zooids. Note polymorphism - gastrozooids (with feeding tentacles) & gonozooids for reproduction [fig 2.3-6] It floats like like boat and Stings like a bee It’s squishy and ghoulish And dangerous to me…. So what is it? Clue - Hydrozoan PHYLUM Cnidaria CLASS Hydrozoa Portuguese Man-O-War is an excellent example of polymorphism. It is a colony of many individuals – again = zooids – modified for different tasks (feeding, floating, reproduction, etc.) This next specimen is on almost EVERY practical exam! PHYLUM Cnidaria CLASS Hydrozoa Calcium-carbonate skeletons of a fire coral.