Gene Encoding Carnitine Dehydratase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection. -

Production of L-Carnitine by Secondary Metabolism of Bacteria Vicente Bernal, Ángel Sevilla, Manuel Cánovas and José L Iborra*
Microbial Cell Factories BioMed Central Review Open Access Production of L-carnitine by secondary metabolism of bacteria Vicente Bernal, Ángel Sevilla, Manuel Cánovas and José L Iborra* Address: Department of Biochemistry and Molecular Biology B and Immunology, Campus of Espinardo, University of Murcia, E-30100, Spain Email: Vicente Bernal - [email protected]; Ángel Sevilla - [email protected]; Manuel Cánovas - [email protected]; José L Iborra* - [email protected] * Corresponding author Published: 2 October 2007 Received: 19 July 2007 Accepted: 2 October 2007 Microbial Cell Factories 2007, 6:31 doi:10.1186/1475-2859-6-31 This article is available from: http://www.microbialcellfactories.com/content/6/1/31 © 2007 Bernal et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract The increasing commercial demand for L-carnitine has led to a multiplication of efforts to improve its production with bacteria. The use of different cell environments, such as growing, resting, permeabilized, dried, osmotically stressed, freely suspended and immobilized cells, to maintain enzymes sufficiently active for L-carnitine production is discussed in the text. The different cell states of enterobacteria, such as Escherichia coli and Proteus sp., which can be used to produce L- carnitine from crotonobetaine or D-carnitine as substrate, are analyzed. Moreover, the combined application of both bioprocess and metabolic engineering has allowed a deeper understanding of the main factors controlling the production process, such as energy depletion and the alteration of the acetyl-CoA/CoA ratio which are coupled to the end of the biotransformation. -

Supplementary Table S1. the Key Enzymes for Autotrophic Growth in C
Electronic Supplementary Material (ESI) for Metallomics. This journal is © The Royal Society of Chemistry 2015 Supplementary Table S1. The key enzymes for autotrophic growth in C. metallidurans Name Rmet-number Spec. Predicted Mass, (kDa) Determined Mass, (kDa) Activity HOS Rmet_1522 to Rmet_1525 28.7 65.2, 25.4, 23.4, 52.8 = 166.8 62.4±1.8, 26.4±1.3, 24.0±0.6, 54.1±2.5, =235±20a HOP Rmet_1297, Rmet_1298, 67.0 69.0 + 38.6 = 108 148±24a CAX Rmet_1500, Rmet_1501 2.82 8 * (13.6 + 52.5) = 529 475±36a PRK Rmet_1512 0.994 8 * 32.4 = 259 256±11a aMean value of masses determined by S300 size exclusion chromatography and sucrose gradient centrifugation. HOS, NAD-reducing soluble hydrogenase; HOP, membrane-bound hydrogenase; CAX, ribulose-bis-phosphate carboxylase/oxygenase; PRK, phosphoribulokinase. Specific activity in U/mg protein. Supplementary Table S2. Genes expressed differently in AE104 compared to CH34 wild typea Operon Region Name Gene Q D Description UP Op1321r Rmet_4594 zntA 1.72 2.88 Q1LEH0 Heavy metal translocating P -type ATPase Op1322f Rmet_4595 czcI2 2.03 2.92 Q1LEG9 Putative uncharacterized protein Op1322f Rmet_4596 czcC2 23.34 5.36 Q1LEG8 Outer membrane efflux protein Op1322f Rmet_4597 czcB2' 15.36 9.68 Q1LEG7 Secretion protein HlyD Op0075f Rmet_0260 - 2.31 2.48 Q1LRT0 Putative transmembrane protein Op0075f Rmet_0261 coxB 2.08 2.49 Q1LRS9 Cytochrome c oxidase subunit 2 Adjacent to CMGI-7 Op0335f Rmet_1171 tnpA 7.03 21.74 Q9F8S6 Transposase (Transposase, IS4 family) CMGI-2 Op0362r Rmet_1251 tnp 4.08 0.61 Q1LNY9 Putative uncharacterized -

B Number Gene Name Strand Orientation Protein Length Mrna
list list sample) short list predicted B number Operon ID Gene name assignment Protein length mRNA present mRNA intensity Gene description Protein detected - Strand orientation Membrane protein detected (total list) detected (long list) membrane sample Proteins detected - detected (short list) # of tryptic peptides # of tryptic peptides # of tryptic peptides # of tryptic peptides # of tryptic peptides Functional category detected (membrane Protein detected - total Protein detected - long b0001 thrL + 21 1344 P 1 0 0 0 0 thr operon leader peptide Metabolism of small molecules 1 b0002 thrA + 820 13624 P 39 P 18 P 18 P 18 P(m) 2 aspartokinase I, homoserine dehydrogenase I Metabolism of small molecules 1 b0003 thrB + 310 6781 P 9 P 3 3 P 3 0 homoserine kinase Metabolism of small molecules 1 b0004 thrC + 428 15039 P 18 P 10 P 11 P 10 0 threonine synthase Metabolism of small molecules 1 b0005 b0005 + 98 432 A 5 0 0 0 0 orf, hypothetical protein Open reading frames 2 b0006 yaaA - 258 1047 P 11 P 1 2 P 1 0 orf, hypothetical protein Open reading frames 3 b0007 yaaJ - 476 342 P 8 0 0 0 0 MP-GenProt-PHD inner membrane transport protein Miscellaneous 4 b0008 talB + 317 20561 P 20 P 13 P 16 P 13 0 transaldolase B Metabolism of small molecules 5 b0009 mog + 195 1296 P 7 0 0 0 0 required for the efficient incorporation of molybdate into molybdoproteins Metabolism of small molecules 6 b0010 yaaH - 188 407 A 2 0 0 0 0 PHD orf, hypothetical protein Open reading frames 7 b0011 b0011 - 237 338 P 13 0 0 0 0 putative oxidoreductase Miscellaneous 8 b0012 htgA -

Viewed and Published Immediately Upon Acceptance Cited in Pubmed and Archived on Pubmed Central Yours — You Keep the Copyright
BMC Bioinformatics BioMed Central Methodology article Open Access Optimization based automated curation of metabolic reconstructions Vinay Satish Kumar1, Madhukar S Dasika2 and Costas D Maranas*2 Address: 1Department of Industrial and Manufacturing Engineering, The Pennsylvania State University, University Park, PA 16802, USA and 2Department of Chemical Engineering, The Pennsylvania State University, University Park, PA 16802, USA Email: Vinay Satish Kumar - [email protected]; Madhukar S Dasika - [email protected]; Costas D Maranas* - [email protected] * Corresponding author Published: 20 June 2007 Received: 14 December 2006 Accepted: 20 June 2007 BMC Bioinformatics 2007, 8:212 doi:10.1186/1471-2105-8-212 This article is available from: http://www.biomedcentral.com/1471-2105/8/212 © 2007 Satish Kumar et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Currently, there exists tens of different microbial and eukaryotic metabolic reconstructions (e.g., Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis) with many more under development. All of these reconstructions are inherently incomplete with some functionalities missing due to the lack of experimental and/or homology information. A key challenge in the automated generation of genome-scale reconstructions is the elucidation of these gaps and the subsequent generation of hypotheses to bridge them. Results: In this work, an optimization based procedure is proposed to identify and eliminate network gaps in these reconstructions. First we identify the metabolites in the metabolic network reconstruction which cannot be produced under any uptake conditions and subsequently we identify the reactions from a customized multi-organism database that restores the connectivity of these metabolites to the parent network using four mechanisms. -

The Analysis of the Intramacrophagic Virulome of Brucella Suis Deciphers the Environment Encountered by the Pathogen Inside the Macrophage Host Cell
The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell Stephan Ko¨ hler*†‡, Vincent Foulongne†§, Safia Ouahrani-Bettache*, Gise` le Bourg§, Jacques Teyssier*, Michel Ramuz§, and Jean-Pierre Liautard* *Institut National de la Sante´et de la Recherche Me´dicale U-431 (INSERM U-431), Universite´Montpellier II, 34095 Montpellier, France; and §INSERM U-431, Faculte´deMe´ decine, Avenue Kennedy, 30900 Nıˆmes,France Edited by Roy Curtiss III, Washington University, St. Louis, MO, and approved September 30, 2002 (received for review July 31, 2002) The pathogen Brucella suis resides and multiplies within a phago- ability to infect animal cells (or protists for Legionella), because cytic vacuole of its host cell, the macrophage. The resulting com- they do not survive for protracted periods of time outside their plex relationship has been investigated by the analysis of the set hosts. These pathogens should therefore rather be considered of genes required for virulence, which we call intramacrophagic mainly as intracellular, facultatively extracellular organisms (4). virulome. Ten thousand two hundred and seventy-two miniTn5 The phagosome, to which they have adapted during evolution, is mutants of B. suis constitutively expressing gfp were screened by their natural niche. fluorescence microscopy for lack of intracellular multiplication in The identification of the whole set of genes involved in the human macrophages. One hundred thirty-one such mutants af- multiplication of Brucella spp. inside the macrophage is a fected in 59 different genes could be isolated, and a function was prerequisite to the understanding of the relation with the ascribed to 53 of them. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Catalysis Science & Technology
Catalysis Science & Technology Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/catalysis Page 1 of 34 Catalysis Science & Technology The selective addition of water Verena Resch a,b and Ulf Hanefeld* a a Gebouw voor Scheikunde, Biokatalyse, Afdeling Biotechnologie, Technische Universiteit Delft, Manuscript Julianalaan 136, 2628BL Delft, Nederland. bOrganische und Bioorganische Chemie, Institut für Chemie, Karl-Franzens-Universität Graz, Heinrichstrasse 28, 8010 Graz, Österreich. Abstract: Water is omnipresent and essential. Yet at the same time it is a rather unreactive Accepted molecule. The direct addition of water to C=C double bonds is therefore a challenge not answered convincingly. In this perspective we critically evaluate the selectivity and the applicability of the different catalytic approaches for water addition reactions, homogeneous, heterogeneous and bio- catalytic. -
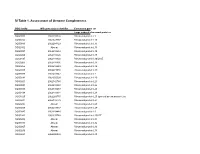
SI Table 1. Assessment of Genome Completeness
SI Table 1. Assessment of Genome Completeness COG family IMG gene object identifier Conserved gene set Large subunit ribosomal proteins COG0081 2062288324 Ribosomal protein L1 COG0244 2062347387 Ribosomal protein L10 COG0080 2062288323 Ribosomal protein L11 COG0102 Absent Ribosomal protein L13 COG0093 2062418832 Ribosomal protein L14 COG0200 2062418826 Ribosomal protein L15 COG0197 2062418838 Ribosomal protein L16/L10E COG0203 2062418836 Ribosomal protein L17 COG0256 2062418829 Ribosomal protein L18 COG0335 2062273558 Ribosomal protein L19 COG0090 2062418842 Ribosomal protein L2 COG0292 2062350539 Ribosomal protein L20 COG0261 2062142780 Ribosomal protein L21 COG0091 2062418840 Ribosomal protein L22 COG0089 2062138283 Ribosomal protein L23 COG0198 2062418834 Ribosomal protein L24 COG1825 2062269715 Ribosomal protein L25 (general stress protein Ctc) COG0211 2062142779 Ribosomal protein L27 COG0227 Absent Ribosomal protein L28 COG0255 2062418837 Ribosomal protein L29 COG0087 2062154483 Ribosomal protein L3 COG1841 2062335748 Ribosomal protein L30/L7E COG0254 Absent Ribosomal protein L31 COG0333 Absent Ribosomal protein L32 COG0267 Absent Ribosomal protein L33 COG0230 Absent Ribosomal protein L34 COG0291 2062350538 Ribosomal protein L35 COG0257 Absent Ribosomal protein L36 COG0088 2062138282 Ribosomal protein L4 COG0094 2062418833 Ribosomal protein L5 COG0097 2062418830 Ribosomal protein L6P/L9E COG0222 2062288326 Ribosomal protein L7/L12 COG0359 2062209880 Ribosomal protein L9 Small subunit ribosomal proteins COG0539 Absent Ribosomal protein -

1471-2164-6-174-S4.PDF (299.1Kb)
Sup_Table_2. Comparison of the whole genomes in Fig. 3A. Segment 1- Conserved in Bm, Bp, and Bt to Bp to Bt Gene Description % length % identity % length % identity BMA0001 chromosomal replication initiator protein DnaA 100 99 100 96 BMA0002 DNA polymerase III, beta subunit 100 100 100 99 BMA0003 DNA gyrase, B subunit 100 100 100 99 BMA0006 carboxymuconolactone decarboxylase family protein 100 98 100 99 BMA0010 hypothetical protein 100 99 100 92 BMA0011 hypothetical protein 100 100 100 91 BMA0014.1 hypothetical protein 100 99 96 94 BMA0018 hypothetical protein 100 99 100 95 BMA0019 FHA domain protein 100 100 100 94 BMA0020 protein kinase domain protein 100 99 100 90 BMA0023 conserved hypothetical protein 100 99 100 90 BMA0024 aldolase, class II 100 98 100 91 BMA0027 polysaccharide biosynthesis family protein 100 100 100 96 BMA0028 glycosyl transferase, group 1 family protein 100 99 100 94 BMA0029 mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase 100 99 100 92 BMA0030 ElaA family protein 100 99 100 90 BMA0032 glycosyl transferase, group 1 family protein 100 99 100 93 BMA0037 sigma-54 dependent transcriptional regulator 100 99 100 97 BMA0039 beta-mannosidase-related protein 100 99 100 91 BMA0040 conserved hypothetical protein 100 100 100 94 BMA0041 conserved hypothetical protein 100 99 100 95 BMA0042 acyl-CoA dehydrogenase domain protein 100 99 100 96 BMA0043 acyl carrier protein, putative 100 100 100 95 BMA0044 conserved hypothetical protein 100 99 100 96 BMA0045 conserved hypothetical protein 100 100 100 98 BMA0046 -

(12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur Et Al
US 2012026.6329A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur et al. (43) Pub. Date: Oct. 18, 2012 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/10 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/24 (2006.01) CI2N 9/02 (2006.01) (75) Inventors: Eric J. Mathur, Carlsbad, CA CI2N 9/06 (2006.01) (US); Cathy Chang, San Marcos, CI2P 2L/02 (2006.01) CA (US) CI2O I/04 (2006.01) CI2N 9/96 (2006.01) (73) Assignee: BP Corporation North America CI2N 5/82 (2006.01) Inc., Houston, TX (US) CI2N 15/53 (2006.01) CI2N IS/54 (2006.01) CI2N 15/57 2006.O1 (22) Filed: Feb. 20, 2012 CI2N IS/60 308: Related U.S. Application Data EN f :08: (62) Division of application No. 1 1/817,403, filed on May AOIH 5/00 (2006.01) 7, 2008, now Pat. No. 8,119,385, filed as application AOIH 5/10 (2006.01) No. PCT/US2006/007642 on Mar. 3, 2006. C07K I4/00 (2006.01) CI2N IS/II (2006.01) (60) Provisional application No. 60/658,984, filed on Mar. AOIH I/06 (2006.01) 4, 2005. CI2N 15/63 (2006.01) Publication Classification (52) U.S. Cl. ................... 800/293; 435/320.1; 435/252.3: 435/325; 435/254.11: 435/254.2:435/348; (51) Int. Cl. 435/419; 435/195; 435/196; 435/198: 435/233; CI2N 15/52 (2006.01) 435/201:435/232; 435/208; 435/227; 435/193; CI2N 15/85 (2006.01) 435/200; 435/189: 435/191: 435/69.1; 435/34; CI2N 5/86 (2006.01) 435/188:536/23.2; 435/468; 800/298; 800/320; CI2N 15/867 (2006.01) 800/317.2: 800/317.4: 800/320.3: 800/306; CI2N 5/864 (2006.01) 800/312 800/320.2: 800/317.3; 800/322; CI2N 5/8 (2006.01) 800/320.1; 530/350, 536/23.1: 800/278; 800/294 CI2N I/2 (2006.01) CI2N 5/10 (2006.01) (57) ABSTRACT CI2N L/15 (2006.01) CI2N I/19 (2006.01) The invention provides polypeptides, including enzymes, CI2N 9/14 (2006.01) structural proteins and binding proteins, polynucleotides CI2N 9/16 (2006.01) encoding these polypeptides, and methods of making and CI2N 9/20 (2006.01) using these polynucleotides and polypeptides. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA