Viewed and Published Immediately Upon Acceptance Cited in Pubmed and Archived on Pubmed Central Yours — You Keep the Copyright
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Cyanide Hydratase from Neurospora Crassa Forms a Helix Which Has a Dimeric Repeat
Appl Microbiol Biotechnol (2009) 82:271–278 DOI 10.1007/s00253-008-1735-4 BIOTECHNOLOGICALLY RELEVANT ENZYMES AND PROTEINS The cyanide hydratase from Neurospora crassa forms a helix which has a dimeric repeat Kyle C. Dent & Brandon W. Weber & Michael J. Benedik & B. Trevor Sewell Received: 2 July 2008 /Revised: 24 September 2008 /Accepted: 25 September 2008 / Published online: 23 October 2008 # Springer-Verlag 2008 Abstract The fungal cyanide hydratases form a functionally Introduction specialized subset of the nitrilases which catalyze the hydrolysis of cyanide to formamide with high specificity. The generation of cyanide containing waste results from These hold great promise for the bioremediation of cyanide cyanide utilization in a variety of applications, spanning wastes. The low resolution (3.0 nm) three-dimensional such diverse industries as gold and silver mining, metal reconstruction of negatively stained recombinant cyanide electroplating, polymer synthesis, and the production of hydratase fibers from the saprophytic fungus Neurospora fine chemicals, pharmaceuticals, dyes, and agricultural crassa by iterative helical real space reconstruction reveals products (Baxter and Cummings 2006). Traditionally, that enzyme fibers display left-handed D1 S5.4 symmetry chemical oxidation or stabilizations are used to treat these with a helical rise of 1.36 nm. This arrangement differs from wastes; however, many groups have been studying the previously characterized microbial nitrilases which demon- potential for bioremediation of such cyanide waste strate a structure built along similar principles but with a (O’Reilly and Turner 2003; Jandhyala et al. 2005). reduced helical twist. The cyanide hydratase assembly is The most promising approach utilizes cyanide degrada- stabilized by two dyadic interactions between dimers across tion by enzymes belonging to the nitrilase family (Pace and the one-start helical groove. -

Cyanide-Degrading Enzymes for Bioremediation A
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Texas A&M University CYANIDE-DEGRADING ENZYMES FOR BIOREMEDIATION A Thesis by LACY JAMEL BASILE Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2008 Major Subject: Microbiology CYANIDE-DEGRADING ENZYMES FOR BIOREMEDIATION A Thesis by LACY JAMEL BASILE Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Approved by: Chair of Committee, Michael Benedik Committee Members, Susan Golden James Hu Wayne Versaw Head of Department, Vincent Cassone August 2008 Major Subject: Microbiology iii ABSTRACT Cyanide-Degrading Enzymes for Bioremediation. (August 2008) Lacy Jamel Basile, B.S., Texas A&M University Chair of Advisory Committee: Dr. Michael Benedik Cyanide-containing waste is an increasingly prevalent problem in today’s society. There are many applications that utilize cyanide, such as gold mining and electroplating, and these processes produce cyanide waste with varying conditions. Remediation of this waste is necessary to prevent contamination of soils and water. While there are a variety of processes being used, bioremediation is potentially a more cost effective alternative. A variety of fungal species are known to degrade cyanide through the action of cyanide hydratases, a specialized subset of nitrilases which hydrolyze cyanide to formamide. Here I report on previously unknown and uncharacterized nitrilases from Neurospora crassa, Gibberella zeae, and Aspergillus nidulans. Recombinant forms of four cyanide hydratases from N. -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection. -

Structures, Functions, and Mechanisms of Filament Forming Enzymes: a Renaissance of Enzyme Filamentation
Structures, Functions, and Mechanisms of Filament Forming Enzymes: A Renaissance of Enzyme Filamentation A Review By Chad K. Park & Nancy C. Horton Department of Molecular and Cellular Biology University of Arizona Tucson, AZ 85721 N. C. Horton ([email protected], ORCID: 0000-0003-2710-8284) C. K. Park ([email protected], ORCID: 0000-0003-1089-9091) Keywords: Enzyme, Regulation, DNA binding, Nuclease, Run-On Oligomerization, self-association 1 Abstract Filament formation by non-cytoskeletal enzymes has been known for decades, yet only relatively recently has its wide-spread role in enzyme regulation and biology come to be appreciated. This comprehensive review summarizes what is known for each enzyme confirmed to form filamentous structures in vitro, and for the many that are known only to form large self-assemblies within cells. For some enzymes, studies describing both the in vitro filamentous structures and cellular self-assembly formation are also known and described. Special attention is paid to the detailed structures of each type of enzyme filament, as well as the roles the structures play in enzyme regulation and in biology. Where it is known or hypothesized, the advantages conferred by enzyme filamentation are reviewed. Finally, the similarities, differences, and comparison to the SgrAI system are also highlighted. 2 Contents INTRODUCTION…………………………………………………………..4 STRUCTURALLY CHARACTERIZED ENZYME FILAMENTS…….5 Acetyl CoA Carboxylase (ACC)……………………………………………………………………5 Phosphofructokinase (PFK)……………………………………………………………………….6 -

Changes in the Sclerotinia Sclerotiorum Transcriptome During Infection of Brassica Napus
Seifbarghi et al. BMC Genomics (2017) 18:266 DOI 10.1186/s12864-017-3642-5 RESEARCHARTICLE Open Access Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus Shirin Seifbarghi1,2, M. Hossein Borhan1, Yangdou Wei2, Cathy Coutu1, Stephen J. Robinson1 and Dwayne D. Hegedus1,3* Abstract Background: Sclerotinia sclerotiorum causes stem rot in Brassica napus, which leads to lodging and severe yield losses. Although recent studies have explored significant progress in the characterization of individual S. sclerotiorum pathogenicity factors, a gap exists in profiling gene expression throughout the course of S. sclerotiorum infection on a host plant. In this study, RNA-Seq analysis was performed with focus on the events occurring through the early (1 h) to the middle (48 h) stages of infection. Results: Transcript analysis revealed the temporal pattern and amplitude of the deployment of genes associated with aspects of pathogenicity or virulence during the course of S. sclerotiorum infection on Brassica napus. These genes were categorized into eight functional groups: hydrolytic enzymes, secondary metabolites, detoxification, signaling, development, secreted effectors, oxalic acid and reactive oxygen species production. The induction patterns of nearly all of these genes agreed with their predicted functions. Principal component analysis delineated gene expression patterns that signified transitions between pathogenic phases, namely host penetration, ramification and necrotic stages, and provided evidence for the occurrence of a brief biotrophic phase soon after host penetration. Conclusions: The current observations support the notion that S. sclerotiorum deploys an array of factors and complex strategies to facilitate host colonization and mitigate host defenses. This investigation provides a broad overview of the sequential expression of virulence/pathogenicity-associated genes during infection of B. -

Production of L-Carnitine by Secondary Metabolism of Bacteria Vicente Bernal, Ángel Sevilla, Manuel Cánovas and José L Iborra*
Microbial Cell Factories BioMed Central Review Open Access Production of L-carnitine by secondary metabolism of bacteria Vicente Bernal, Ángel Sevilla, Manuel Cánovas and José L Iborra* Address: Department of Biochemistry and Molecular Biology B and Immunology, Campus of Espinardo, University of Murcia, E-30100, Spain Email: Vicente Bernal - [email protected]; Ángel Sevilla - [email protected]; Manuel Cánovas - [email protected]; José L Iborra* - [email protected] * Corresponding author Published: 2 October 2007 Received: 19 July 2007 Accepted: 2 October 2007 Microbial Cell Factories 2007, 6:31 doi:10.1186/1475-2859-6-31 This article is available from: http://www.microbialcellfactories.com/content/6/1/31 © 2007 Bernal et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract The increasing commercial demand for L-carnitine has led to a multiplication of efforts to improve its production with bacteria. The use of different cell environments, such as growing, resting, permeabilized, dried, osmotically stressed, freely suspended and immobilized cells, to maintain enzymes sufficiently active for L-carnitine production is discussed in the text. The different cell states of enterobacteria, such as Escherichia coli and Proteus sp., which can be used to produce L- carnitine from crotonobetaine or D-carnitine as substrate, are analyzed. Moreover, the combined application of both bioprocess and metabolic engineering has allowed a deeper understanding of the main factors controlling the production process, such as energy depletion and the alteration of the acetyl-CoA/CoA ratio which are coupled to the end of the biotransformation. -

Electron Transport Phosphorylation in Rumen Butyrivibrios: Unprecedented ATP Yield for Glucose Fermentation to Butyrate
HYPOTHESIS AND THEORY published: 24 June 2015 doi: 10.3389/fmicb.2015.00622 Electron transport phosphorylation in rumen butyrivibrios: unprecedented ATP yield for glucose fermentation to butyrate Timothy J. Hackmann1 and Jeffrey L. Firkins2* 1 Department of Animal Sciences, University of Florida, Gainesville, FL, USA, 2 Department of Animal Sciences, The Ohio State University, Columbus, OH, USA From a genomic analysis of rumen butyrivibrios (Butyrivibrio and Pseudobutyrivibrio sp.), we have re-evaluated the contribution of electron transport phosphorylation (ETP) to ATP formation in this group. This group is unique in that most (76%) genomes were predicted to possess genes for both Ech and Rnf transmembrane ion pumps. These pumps act in concert with the NifJ and Bcd-Etf to form a electrochemical potential (μH+ and μNa+), which drives ATP synthesis by ETP. Of the 62 total butyrivibrio genomes currently available from the Hungate 1000 project, all 62 were predicted to Edited by: possess NifJ, which reduces oxidized ferredoxin (Fdox) during pyruvate conversion to Emilio M. Ungerfeld, Instituto de Investigaciones acetyl-CoA. All 62 possessed all subunits of Bcd-Etf, which reduces Fdox and oxidizes Agropecuarias, Chile reduced NAD during crotonyl-CoA reduction. Additionally, 61 genomes possessed all Reviewed by: subunits of the Rnf, which generates μH+ or μNa+ from oxidation of reduced Fd Wolfgang Buckel, (Fdred) and reduction of oxidized NAD. Further, 47 genomes possessed all six subunits Philipps-Universität Marburg, + Germany of the Ech, which generates μH from oxidation of Fdred. For glucose fermentation Robert J. Wallace, to butyrate and H2, the electrochemical potential established should drive synthesis University of Aberdeen, UK of ∼1.5 ATP by the F0F1-ATP synthase (possessed by all 62 genomes). -

Supplementary Informations SI2. Supplementary Table 1
Supplementary Informations SI2. Supplementary Table 1. M9, soil, and rhizosphere media composition. LB in Compound Name Exchange Reaction LB in soil LBin M9 rhizosphere H2O EX_cpd00001_e0 -15 -15 -10 O2 EX_cpd00007_e0 -15 -15 -10 Phosphate EX_cpd00009_e0 -15 -15 -10 CO2 EX_cpd00011_e0 -15 -15 0 Ammonia EX_cpd00013_e0 -7.5 -7.5 -10 L-glutamate EX_cpd00023_e0 0 -0.0283302 0 D-glucose EX_cpd00027_e0 -0.61972444 -0.04098397 0 Mn2 EX_cpd00030_e0 -15 -15 -10 Glycine EX_cpd00033_e0 -0.0068175 -0.00693094 0 Zn2 EX_cpd00034_e0 -15 -15 -10 L-alanine EX_cpd00035_e0 -0.02780553 -0.00823049 0 Succinate EX_cpd00036_e0 -0.0056245 -0.12240603 0 L-lysine EX_cpd00039_e0 0 -10 0 L-aspartate EX_cpd00041_e0 0 -0.03205557 0 Sulfate EX_cpd00048_e0 -15 -15 -10 L-arginine EX_cpd00051_e0 -0.0068175 -0.00948672 0 L-serine EX_cpd00054_e0 0 -0.01004986 0 Cu2+ EX_cpd00058_e0 -15 -15 -10 Ca2+ EX_cpd00063_e0 -15 -100 -10 L-ornithine EX_cpd00064_e0 -0.0068175 -0.00831712 0 H+ EX_cpd00067_e0 -15 -15 -10 L-tyrosine EX_cpd00069_e0 -0.0068175 -0.00233919 0 Sucrose EX_cpd00076_e0 0 -0.02049199 0 L-cysteine EX_cpd00084_e0 -0.0068175 0 0 Cl- EX_cpd00099_e0 -15 -15 -10 Glycerol EX_cpd00100_e0 0 0 -10 Biotin EX_cpd00104_e0 -15 -15 0 D-ribose EX_cpd00105_e0 -0.01862144 0 0 L-leucine EX_cpd00107_e0 -0.03596182 -0.00303228 0 D-galactose EX_cpd00108_e0 -0.25290619 -0.18317325 0 L-histidine EX_cpd00119_e0 -0.0068175 -0.00506825 0 L-proline EX_cpd00129_e0 -0.01102953 0 0 L-malate EX_cpd00130_e0 -0.03649016 -0.79413596 0 D-mannose EX_cpd00138_e0 -0.2540567 -0.05436649 0 Co2 EX_cpd00149_e0 -
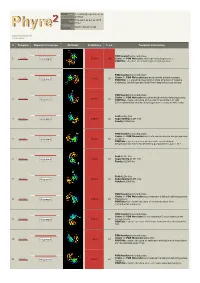
Phyre 2 Results for P25553
Email [email protected] Description P25553 Thu Jan 5 11:42:11 GMT Date 2012 Unique Job 5024f4b9e5342484 ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:oxidoreductase 1 c2hg2A_ Alignment 100.0 100 Chain: A: PDB Molecule:aldehyde dehydrogenase a; PDBTitle: structure of lactaldehyde dehydrogenase PDB header:oxidoreductase Chain: B: PDB Molecule:betaine aldehyde dehydrogenase; 2 c3ed6B_ 100.0 36 Alignment PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus PDB header:oxidoreductase Chain: A: PDB Molecule:formyltetrahydrofolate dehydrogenase; 3 c2o2qA_ 100.0 33 Alignment PDBTitle: crystal structure of the c-terminal domain of rat2 10'formyltetrahydrofolate dehydrogenase in complex with nadp Fold:ALDH-like 4 d1a4sa_ Alignment 100.0 35 Superfamily:ALDH-like Family:ALDH-like PDB header:oxidoreductase Chain: H: PDB Molecule:succinate-semialdehyde dehydrogenase 5 c3ifgH_ Alignment 100.0 34 (nadp+); PDBTitle: crystal structure of succinate-semialdehyde dehydrogenase from2 burkholderia pseudomallei, part 1 of 2 Fold:ALDH-like 6 d1bxsa_ Alignment 100.0 33 Superfamily:ALDH-like Family:ALDH-like Fold:ALDH-like 7 d1o9ja_ Alignment 100.0 33 Superfamily:ALDH-like Family:ALDH-like PDB header:oxidoreductase Chain: A: PDB Molecule:succinate-semialdehyde dehydrogenase 8 c3rh9A_ Alignment 100.0 35 (nad(p)(+)); PDBTitle: the crystal structure of oxidoreductase from marinobacter aquaeolei PDB header:oxidoreductase Chain: B: PDB Molecule:5-carboxymethyl-2-hydroxymuconate 9 c2d4eB_ Alignment 100.0 32 semialdehyde PDBTitle: crystal structure of the hpcc from thermus thermophilus hb8 PDB header:oxidoreductase Chain: G: PDB Molecule:antiquitin; 10 c2jg7G_ 100.0 28 Alignment PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity PDB header:oxidoreductase Chain: C: PDB Molecule:succinate-semialdehyde dehydrogenase 11 c3jz4C_ 100.0 39 Alignment [nadp+]; PDBTitle: crystal structure of e. -

Investigation of Iron's Role in Alginate Production And
INVESTIGATION OF IRON’S ROLE IN ALGINATE PRODUCTION AND MUCOIDY BY PSEUDOMONAS AERUGINOSA by JACINTA ROSE SCHRAUFNAGEL B.S., University of Colorado Denver, 2003 M.S., University of Colorado Denver, 2006 A thesis submitted to the Faculty of the Graduate School of the University of Colorado in partial fulfillment of the requirements for the degree of Doctor of Philosophy Molecular Biology Program 2013 This thesis for the Doctor of Philosophy degree by Jacinta Rose Schraufnagel has been approved for the Molecular Biology Program by David Barton, Chair Michael Vasil, Advisor Mair Churchill Jerry Schaack Michael Schurr Martin Voskuil Date 10/30/13 ii Schraufnagel, Jacinta R. (Ph.D., Molecular Biology) Investigation of Iron’s Role in Alginate Production and Mucoidy by Pseudomonas aeruginosa. Thesis directed by Professor Michael Vasil. ABSTRACT Iron, an essential nutrient, influences the production of major virulence factors and the development of structured non-alginate biofilms in Pseudomonas aeruginosa. Fur, pyoverdine and various concentrations of iron sources have all been shown to affect non-alginate biofilm formation. Remarkably however, the role of iron in alginate biofilm formation and mucoidy is unknown. Because of alginate’s association with biofilms in chronic cystic fibrosis (CF) lung infections, we examined the influence of iron on alginate production and mucoidy in P. aeruginosa. Herein, we identify and characterize a novel iron-regulated protein that affects the transcription of alginate biosynthesis genes. Further, contrary to what is known about non-alginate biofilms, we discovered that biologically relevant concentrations of iron (10 µM – 300 µM), in various forms (e.g. FeCl3, Ferric Citrate, Heme), led to the decreased expression of alginate biosynthesis genes and the dispersion of alginate biofilms in mucoid P. -

Table 4. V. Cholerae Flexgene ORF Collection
Table 4. V. cholerae FLEXGene ORF collection Reference Clone protein PlasmID clone GenBank Locus tag Symbol accession identifier FLEX clone name accession Product name VC0001 NP_062585 VcCD00019918 FLH200476.01F DQ772770 hypothetical protein VC0002 mioC NP_062586 VcCD00019938 FLH200506.01F DQ772771 mioC protein VC0003 thdF NP_062587 VcCD00019958 FLH200531.01F DQ772772 thiophene and furan oxidation protein ThdF VC0004 yidC NP_062588 VcCD00019970 FLH200545.01F DQ772773 inner membrane protein, 60 kDa VC0005 NP_062589 VcCD00061243 FLH236482.01F DQ899316 conserved hypothetical protein VC0006 rnpA NP_062590 VcCD00025697 FLH214799.01F DQ772774 ribonuclease P protein component VC0007 rpmH NP_062591 VcCD00061229 FLH236450.01F DQ899317 ribosomal protein L34 VC0008 NP_062592 VcCD00019917 FLH200475.01F DQ772775 amino acid ABC transporter, ATP-binding protein VC0009 NP_062593 VcCD00019966 FLH200540.01F DQ772776 amino acid ABC transproter, permease protein VC0010 NP_062594 VcCD00019152 FLH199275.01F DQ772777 amino acid ABC transporter, periplasmic amino acid-binding portion VC0011 NP_062595 VcCD00019151 FLH199274.01F DQ772778 hypothetical protein VC0012 dnaA NP_062596 VcCD00017363 FLH174286.01F DQ772779 chromosomal DNA replication initiator DnaA VC0013 dnaN NP_062597 VcCD00017316 FLH174063.01F DQ772780 DNA polymerase III, beta chain VC0014 recF NP_062598 VcCD00019182 FLH199319.01F DQ772781 recF protein VC0015 gyrB NP_062599 VcCD00025458 FLH174642.01F DQ772782 DNA gyrase, subunit B VC0016 NP_229675 VcCD00019198 FLH199346.01F DQ772783 hypothetical protein -

Diversity of Antisense and Other Non-Coding Rnas in Archaea Revealed by Comparative Small RNA Sequencing in Four Pyrobaculum Species
Lawrence Berkeley National Laboratory Recent Work Title Diversity of Antisense and Other Non-Coding RNAs in Archaea Revealed by Comparative Small RNA Sequencing in Four Pyrobaculum Species. Permalink https://escholarship.org/uc/item/94r6n6t1 Journal Frontiers in microbiology, 3(JUL) ISSN 1664-302X Authors Bernick, David L Dennis, Patrick P Lui, Lauren M et al. Publication Date 2012 DOI 10.3389/fmicb.2012.00231 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ORIGINAL RESEARCH ARTICLE published: 02 July 2012 doi: 10.3389/fmicb.2012.00231 Diversity of antisense and other non-coding RNAs in archaea revealed by comparative small RNA sequencing in four Pyrobaculum species David L. Bernick 1, Patrick P.Dennis 2, Lauren M. Lui 1 andTodd M. Lowe 1* 1 Department of Biomolecular Engineering, University of California, Santa Cruz, CA, USA 2 Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA Edited by: A great diversity of small, non-coding RNA (ncRNA) molecules with roles in gene regula- Frank T. Robb, University of California, tion and RNA processing have been intensely studied in eukaryotic and bacterial model USA organisms, yet our knowledge of possible parallel roles for small RNAs (sRNA) in archaea Reviewed by: Mircea Podar, Oak Ridge National is limited. We employed RNA-seq to identify novel sRNA across multiple species of the Laboratory, USA hyperthermophilic genus Pyrobaculum, known for unusual RNA gene characteristics. By Imke Schroeder, University of comparing transcriptional data collected in parallel among four species, we were able to California Los Angeles, USA identify conserved RNA genes fitting into known and novel families.