Supplementary Table 2: Energy Metabolism Related Genes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Airway Epithelium Undergoes Metabolic Reprogramming in Individuals at High Risk for Lung Cancer
The airway epithelium undergoes metabolic reprogramming in individuals at high risk for lung cancer S.M. Jamshedur Rahman, … , Jamey D. Young, Pierre P. Massion JCI Insight. 2016;1(19):e88814. https://doi.org/10.1172/jci.insight.88814. Research Article Metabolism Oncology The molecular determinants of lung cancer risk remain largely unknown. Airway epithelial cells are prone to assault by risk factors and are considered to be the primary cell type involved in the field of cancerization. To investigate risk- associated changes in the bronchial epithelium proteome that may offer new insights into the molecular pathogenesis of lung cancer, proteins were identified in the airway epithelial cells of bronchial brushing specimens from risk-stratified individuals by shotgun proteomics. Differential expression of selected proteins was validated by parallel reaction monitoring mass spectrometry in an independent set of individual bronchial brushings. We identified 2,869 proteins, of which 312 proteins demonstrated a trend in expression. Pathway analysis revealed enrichment of carbohydrate metabolic enzymes in high-risk individuals. Glucose consumption and lactate production were increased in human bronchial epithelial BEAS2B cells treated with cigarette smoke condensate for 7 months. Increased lipid biosynthetic capacity and 13 net reductive carboxylation were revealed by metabolic flux analyses of [U- C5] glutamine in this in vitro model, suggesting profound metabolic reprogramming in the airway epithelium of high-risk individuals. These results provide a rationale for the development of potentially new chemopreventive strategies and selection of patients for surveillance programs. Find the latest version: https://jci.me/88814/pdf RESEARCH ARTICLE The airway epithelium undergoes metabolic reprogramming in individuals at high risk for lung cancer S.M. -

Class-I and Class-II Fumarases Are a Paradigm of the Recruitment Of
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.232652; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Class-I and Class-II fumarases are a paradigm of the recruitment of 2 metabolites and metabolic enzymes for signalling of the DNA Damage 3 Response during evolution. 4 5 Yardena Silas 1, 2, Esti Singer 1, Norbert Lehming 2 and Ophry Pines 1, 2* 6 1. Department of Microbiology and Molecular Genetics, IMRIC, Faculty of Medicine, 7 Hebrew University, Jerusalem, Israel 8 2. CREATE‑NUS‑HUJ Program and the Department of Microbiology and Immunology, 9 Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 10 Singapore. 11 12 [email protected] 13 [email protected] 14 [email protected] 15 [email protected] 16 17 18 19 20 21 22 23 24 25 26 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.232652; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 27 Abstract 28 Class-II fumarase (Fumarate Hydratase, FH) and its metabolic intermediates are essential 29 components in the DNA damage response (DDR) in eukaryotic cells (human and yeast) and 30 in the prokaryote Bacillus subtilis. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Adaptation to Hif1α Deletion in Hypoxic Cancer Cells by Upregulation of GLUT14 and Creatine Metabolism
Published OnlineFirst March 18, 2019; DOI: 10.1158/1541-7786.MCR-18-0315 Metabolism Molecular Cancer Research Adaptation to HIF1a Deletion in Hypoxic Cancer Cells by Upregulation of GLUT14 and Creatine Metabolism Alessandro Valli1,2, Matteo Morotti1, Christos E. Zois1, Patrick K. Albers3, Tomoyoshi Soga4, Katharina Feldinger1, Roman Fischer2, Martin Frejno2, Alan McIntyre1, Esther Bridges1, Syed Haider1, Francesca M. Buffa1, Dilair Baban3, Miguel Rodriguez5,6, Oscar Yanes5,6, Hannah J. Whittington7, Hannah A. Lake7, Sevasti Zervou7, Craig A. Lygate7, Benedikt M. Kessler2, and Adrian L. Harris1 Abstract Hypoxia-inducible factor 1a is a key regulator of the than phosphofructokinase. Furthermore, glucose uptake hypoxia response in normal and cancer tissues. It is well could be maintained in hypoxia through upregulation of recognized to regulate glycolysis and is a target for therapy. GLUT14, not previously recognized in this role. Finally, However, how tumor cells adapt to grow in the absence of there was a marked adaptation and change of phospho- HIF1a is poorly understood and an important concept to creatine energy pathways, which made the cells susceptible understand for developing targeted therapies is the flexi- to inhibition of creatine metabolism in hypoxic condi- bility of the metabolic response to hypoxia via alternative tions. Overall, our studies show a complex adaptation to pathways. We analyzed pathways that allow cells to survive hypoxia that can bypass HIF1a, but it is targetable and it hypoxic stress in the absence of HIF1a,usingtheHCT116 provides new insight into the key metabolic pathways colon cancer cell line with deleted HIF1a versus control. involved in cancer growth. Spheroids were used to provide a 3D model of metabolic gradients. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Exploring the Non-Canonical Functions of Metabolic Enzymes Peiwei Huangyang1,2 and M
© 2018. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2018) 11, dmm033365. doi:10.1242/dmm.033365 REVIEW SPECIAL COLLECTION: CANCER METABOLISM Hidden features: exploring the non-canonical functions of metabolic enzymes Peiwei Huangyang1,2 and M. Celeste Simon1,3,* ABSTRACT A key finding from studies of metabolic enzymes is the existence The study of cellular metabolism has been rigorously revisited over the of mechanistic links between their nuclear localization and the past decade, especially in the field of cancer research, revealing new regulation of transcription. By modulating gene expression, insights that expand our understanding of malignancy. Among these metabolic enzymes themselves facilitate adaptation to rapidly insights isthe discovery that various metabolic enzymes have surprising changing environments. Furthermore, they can directly shape a ’ activities outside of their established metabolic roles, including in cell s epigenetic landscape (Kaelin and McKnight, 2013). the regulation of gene expression, DNA damage repair, cell cycle Strikingly, several metabolic enzymes exert completely distinct progression and apoptosis. Many of these newly identified functions are functions in different cellular compartments. Nuclear fructose activated in response to growth factor signaling, nutrient and oxygen bisphosphate aldolase, for example, directly interacts with RNA ́ availability, and external stress. As such, multifaceted enzymes directly polymerase III to control transcription (Ciesla et al., 2014), -

PYGL Rabbit Polyclonal Antibody
PYGL Rabbit Polyclonal Antibody CAB6710 Product Information Protein Background Size: This gene encodes a homodimeric protein that catalyses the cleavage of alpha-1, 4-glucosidic bonds to release glucose-1-phosphate from liver glycogen stores. This protein switches from 20uL, 50uL, 100uL, 200uL inactive phosphorylase B to active phosphorylase A by phosphorylation of serine residue 15. Activity of this enzyme is further regulated by multiple allosteric effectors and hormonal Observed MW: controls. Humans have three glycogen phosphorylase genes that encode distinct isozymes that 110kDa are primarily expressed in liver, brain and muscle, respectively. The liver isozyme serves the glycemic demands of the body in general while the brain and muscle isozymes supply just Calculated MW: those tissues. In glycogen storage disease type VI, also known as Hers disease, mutations in liver glycogen phosphorylase inhibit the conversion of glycogen to glucose and results in 93kDa/97kDa moderate hypoglycemia, mild ketosis, growth retardation and hepatomegaly. Alternative splicing results in multiple transcript variants encoding different isoforms. Applications: WB IF IP Immunogen information Reactivity: Gene ID: 5836 Human, Mouse, Rat Uniprot P06737 Antibody Information Recommended dilutions: Synonyms: WB 1:500 - 1:2000 IF 1:50 - PYGL; GSD6 1:200 IP 1:50 - 1:200 Source: Rabbit Immunogen: Recombinant fusion protein containing a sequence corresponding Isotype: to amino acids 1-280 of human PYGL (NP_002854.3). IgG Storage: Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. Purification: Affinity purification Copyright © 2021 Assay Genie [email protected] www.assaygenie.com Product Images Western blot analysis of extracts of various cell lines, using PYGL antibody (CAB6710) at 1:1000 dilution. -

Histone Methyltransferase Smyd1 Regulates Mitochondrial Energetics in the Heart
Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart Junco S. Warrena,b,1, Christopher M. Tracya, Mickey R. Millera, Aman Makajua, Marta W. Szulika, Shin-ichi Okac, Tatiana N. Yuzyukd,e, James E. Coxf,g, Anil Kumarh, Bucky K. Loziere, Li Wanga, June García Llanaa, Amira D. Sabrya, Keiko M. Cawleya, Dane W. Bartonb, Yong Hwan Hani, Sihem Boudinaj, Oliver Fiehnk,l, Haley O. Tuckerm,n, Alexey V. Zaitseva,o, and Sarah Franklina,b,g aNora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, UT 84112; bDepartment of Internal Medicine, University of Utah School of Medicine, Salt Lake City, UT 84102; cDepartment of Cell Biology and Molecular Medicine, Rutgers New Jersey Medical School, Newark, NJ 07103; dDepartment of Pathology, University of Utah School of Medicine, Salt Lake City, UT 84103; eAssociated Regional and University Pathologists, Inc. Laboratories, Salt Lake City, UT 84108; fMetabolomics Core Research Facility, University of Utah, Salt Lake City, UT 84112; gDepartment of Biochemistry, University of Utah, Salt Lake City, UT 84112; hMetabolic Phenotyping Core Facility, University of Utah, Salt Lake City, UT 84112; iMasonic Cancer Center, University of Minnesota, Minneapolis, MN 55455; jDepartment of Nutrition and Integrative Physiology, University of Utah, Salt Lake City, UT 84112; kGenome Center-Metabolomics, University of California, Davis, CA 95616; lBiochemistry Department, King Abdulaziz University, 21589 Jeddah, Saudi Arabia; mDepartment of Molecular Biosciences, University of Texas at Austin, Austin, TX 78712; nThe Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712; and oDepartment of Bioengineering, University of Utah, Salt Lake City, UT 84112 Edited by Gerald I. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Appendix Table A.2.3.1 Full Table of All Chicken Proteins and Human Orthologs Pool Accession Human Human Protein Human Product Cell Angios Log2( Endo Gene Comp
Appendix table A.2.3.1 Full table of all chicken proteins and human orthologs Pool Accession Human Human Protein Human Product Cell AngioS log2( Endo Gene comp. core FC) Specific CIKL F1NWM6 KDR NP_002244 kinase insert domain receptor (a type III receptor tyrosine M 94 4 kinase) CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1N871 FLT4 NP_891555 fms-related tyrosine kinase 4 M 86 -1.71 CWT O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CIKL O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CWT Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL F1NFQ9 TEK NP_000450 TEK tyrosine kinase, endothelial M 77 7.3 specific CWT Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CIKL Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CWT F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CIKL F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CWT E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CIKL E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CWT O93419 COL18A1 NP_569712 collagen, type XVIII, alpha 1 E 70 -2.13 CIKL O93419 -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -
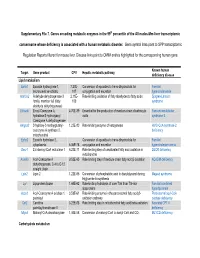
Supplementary File 7. Genes Encoding Metabolic Enzymes in The
Supplementary File 7. Genes encoding metabolic enzymes in the 99th percentile of the All nodes-Mm-liver transcriptomic consensome whose deficiency is associated with a human metabolic disorder. Gene symbol links point to SPP transcriptomic Regulation Reports filtered for mouse liver. Disease links point to OMIM entries highlighted for the corresponding human gene. Known human Target Gene product CPV Hepatic metabolic pathway deficiency disease Lipid metabolism Ephx1 Epoxide hydroxylase 1, 7.27E- Conversion of epoxides to trans-dihydrodiols for Familial microsomal xenobiotic 107 conjugation and excretion hypercholanemia Aldh3a2 Aldehyde dehydrogenase 3 2.11E- Rate-limiting oxidation of fatty aldehydes to fatty acids Sjogren-Larsson family, member A2 (fatty 103 syndrome aldehyde dehydrogenase) Ehhadh Enoyl-Coenzyme A, 4.70E-89 Essential for the production of medium-chain dicarboxylic Fanconi renotubular hydratase/3-hydroxyacyl acids syndrome 3 Coenzyme A dehydrogenase Hmgcs2 3-hydroxy-3-methylglutaryl- 1.23E-80 Rate-limiting enzyme of ketogenesis HMG-CoA synthase-2 coenzyme A synthase 2, deficiency mitochondrial Ephx2 Epoxide hydrolase 2, Conversion of epoxides to trans-dihydrodiols for Familial cytoplasmic 6.06E-78 conjugation and excretion hypercholesterolemia Decr1 2,4-dienoyl-CoA reductase 1 6.23E-71 Rate-limiting step of unsaturated fatty acid oxidation in DECR deficiency mitochondria Acadm Acyl-Coenzyme A 9.52E-65 Rate limiting step of medium-chain fatty acid β-oxidation ACADM deficiency dehydrogenase, C-4 to C-12 straight chain Lpin2