Certificate of Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Aminophylline Catalog Number A1755 Storage
Aminophylline Catalog Number A1755 Storage Temperature –20 °C Replacement for Catalog Number 216895 CAS RN 317-34-0 Storage/Stability Synonyms: theophylline hemiethylenediamine complex; Aminophylline should be kept tightly closed to prevent 3,7-dihydro-1,3-demethyl-1H-purine-2,6-dione CO2 absorption from the atmosphere, which leads to compound with 1,2-ethanediamine (2:1); formation of theophylline and decreased solubility in 1,2 3 (theophylline)2 • ethylenediamine aqueous solutions. Stock solutions should be protected from light and prevented from contact with Product Description metals.2 Molecular Formula: C7H8N4O2 ·1/2 (C2H8N2) Molecular Weight: 210.3 References 1. The Merck Index, 12th ed., Entry# 485. Aminophylline is a xanthine derivative which is a 2. Martindale: The Extra Pharmacopoeia, 31st ed., combination of theophylline and ethylenediamine that is Reynolds, J. E. F., ed., Royal Pharmaceutical more water soluble than theophylline alone. Society (London, England: 1996), pp. 1651-1652. Aminophylline has been widely used as an inhibitor of 3. Data for Biochemical Research, 3rd ed., Dawson, cAMP phosphodiesterase.3 R. M. C., et al., Oxford University Press (New York, NY: 1986), pp. 316-317. Aminophylline has been shown to limit 4. Pelech, S. L., et al., cAMP analogues inhibit phosphatidylcholine biosynthesis in cultured rat phosphatidylcholine biosynthesis in cultured rat hepatocytes.4 It has been used in studies of acute hepatocytes. J. Biol. Chem., 256(16), 8283-8286 hypoxemia in newborn and older guinea pigs.5 The (1981). effect of various xanthine derivatives, including 5. Crisanti, K. C., and Fewell, J. E., Aminophylline aminophylline, on activation of the cystic fibrosis alters the core temperature response to acute transmembrane conductance regulator (CFTR) chloride hypoxemia in newborn and older guinea pigs. -

Download Product Insert (PDF)
PRODUCT INFORMATION Proxyphylline Item No. 20937 CAS Registry No.: 603-00-9 Formal Name: 3,7-dihydro-7-(2-hydroxypropyl)-1,3- N dimethyl-1H-purine-2,6-dione O N Synonym: NSC 163343 C H N O MF: 10 14 4 3 N FW: 238.2 N Purity: ≥98% O UV/Vis.: λmax: 273, 324 nm Supplied as: A crystalline solid OH Storage: -20°C Stability: As supplied, 2 years from the QC date provided on the Certificate of Analysis, when stored properly Laboratory Procedures Proxyphylline is supplied as a crystalline solid. A stock solution may be made by dissolving the proxyphylline in the solvent of choice. Proxyphylline is soluble in organic solvents such as ethanol, DMSO, and dimethyl formamide (DMF), which should be purged with an inert gas. The solubility of proxyphylline in ethanol is approximately 1 mg/ml and approximately 10 mg/ml in DMSO and DMF. Further dilutions of the stock solution into aqueous buffers or isotonic saline should be made prior to performing biological experiments. Ensure that the residual amount of organic solvent is insignificant, since organic solvents may have physiological effects at low concentrations. Organic solvent-free aqueous solutions of proxyphylline can be prepared by directly dissolving the crystalline solid in aqueous buffers. The solubility of proxyphylline in PBS, pH 7.2, is approximately 1 mg/ml. We do not recommend storing the aqueous solution for more than one day. Description Proxyphylline is a methylxanthine derivative that has bronchodilatory actions.1 It has also been reported 2 to have vasodilatory and cardiac stimulatory effects. -

The Xanthines (Theobromine and Aminophyllin)
effect of this might conceivably escape detection in THE XANTHINES (THEOBROMINE AND individuals engaged in very heavy work. From this AMINOPHYLLINE) IN THE TREAT- point of view our subjects were particularly favorable for this since of as MENT OF CARDIAC PAIN study, most them, shown in the table, were not engaged in any occupation. HARRY M.D. GOLD, GLYCERYL TRINITRATE TEST NATHANIEL T. M.D. KWIT, Early in the course of the study it was believed AND desirable to HAROLD M.D. restrict the selection of patients to those OTTO, who could establish their qualifications for service in NEW YORK such a study as this by their ability to distinguish An endeavor was made in this study to secure evi- between the efficacy of glyceryl trinitrate taken under dence on the question of whether the xanthines relieve the tongue and a soluble placebo tablet taken in the cardiac pain. same manner for relief during attacks of pain. The SELECTION OF PATIENTS discovery of several patients who found the two equally effective those who had suffered an of The were 100 ambulant in attend- among attack subjects patients thrombosis and were to thoracic ance at the cardiac in whom the of coronary subject pain clinic, diagnosis on effort led us to abandon this restriction. arteriosclerotic heart disease with cardiac pain was made, in accordance with the nomenclature and criteria The results obtained in sixty patients in whom the the New York Heart Association.1 glyceryl trinitrate test was made are of some interest. adopted by These received trinitrate were selected from a total case load of patients glyceryl tablets, %0o They approxi- or cr 0.4 which were mately 700 patients, representing an average sample /4so grain (0.6 mg.), they of the cardiac clinic several racial directed to take under the tongue at the onset of an population, comprising attack of In of these of groups, both native and born. -

Chocolate, Theobromine, Dogs, and Other Great Stuff
Nancy Lowry, Professor of Chemistry, Hampshire College, Amherst, MA [email protected] Chocolate, Theobromine, Dogs, and Other Great Stuff. Chocolate is now considered a health food, according to many news reports. It provides a goodly dose of antioxidants, prolongs the lives of Dutch men, contains compounds that chemically echo tetrahydocannabinoid and encourage feelings of love, and it even “may halve the risk of dying,” according to a recent headline in the New Scientist. On the other hand, if chocolate is included in the diet in therapeutic doses, it will also most assuredly lead to obesity. Furthermore, the amounts of anandamide (the THC mimic) and phenylethylamine (the so-called “love” compound) are present in chocolate in very, very low amounts. And finally, we all have a 100% chance of dying at some time, so a headline that talks about cutting our chance of dying in half makes no sense. Nevertheless, chocolate is great stuff. It comes in many varieties. One end of the spectrum is bitter baking chocolate; adding sugar provides chocolate of various degrees of sweetness. Adding milk finally brings us to milk chocolate, which many people consider barely makes it over the line into chocolate. White chocolate is only cocoa butter fat, and really isn’t chocolate at all. Over 600 different molecules contribute to the taste of chocolate. Many people talk about the caffeine in chocolate, but there is relatively very little caffeine in chocolate; the compound that particularly characterizes chocolate is theobromine, a very close relative of caffeine. There is six to ten times more theobromine in chocolate than caffeine. -

Soy Isoflavone Genistein Inhibits an Axillary Osmidrosis Risk Factor ABCC11: in Vitro Screening and Fractional Approach for ABCC11-Inhibitory Activities in Plant Extracts and Dietary
nutrients Article Soy Isoflavone Genistein Inhibits an Axillary Osmidrosis Risk Factor ABCC11: In Vitro Screening and Fractional Approach for ABCC11-Inhibitory Activities in Plant Extracts and Dietary Flavonoids 1,2, 2, , 1 1 1 Hiroki Saito y, Yu Toyoda * y , Hiroshi Hirata , Ami Ota-Kontani , Youichi Tsuchiya , Tappei Takada 2 and Hiroshi Suzuki 2 1 Frontier Laboratories for Value Creation, Sapporo Holdings Ltd., 10 Okatome, Yaizu, Shizuoka 425-0013, Japan; [email protected] (H.S.); [email protected] (H.H.); [email protected] (A.O.-K.); [email protected] (Y.T.) 2 Department of Pharmacy, The University of Tokyo Hospital, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; [email protected] (T.T.); [email protected] (H.S.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 2 July 2020; Accepted: 12 August 2020; Published: 14 August 2020 Abstract: Axillary osmidrosis (AO) is a common chronic skin condition characterized by unpleasant body odors emanating from the armpits, and its aetiology is not fully understood. AO can seriously impair the psychosocial well-being of the affected individuals; however, no causal therapy has been established for it other than surgical treatment. Recent studies have revealed that human ATP-binding cassette transporter C11 (ABCC11) is an AO risk factor when it is expressed in the axillary apocrine glands—the sources of the offensive odors. Hence, identifying safe ways to inhibit ABCC11 may offer a breakthrough in treating AO. We herein screened for ABCC11-inhibitory activities in 34 natural products derived from plants cultivated for human consumption using an in vitro assay system to measure the ABCC11-mediated transport of radiolabeled dehydroepiandrosterone sulfate (DHEA-S—an ABCC11 substrate). -

Different Approaches to Evaluate Tannin Content and Structure of Selected Plant Extracts – Review and New Aspects M
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by JKI Open Journal Systems (Julius Kühn-Institut) Journal of Applied Botany and Food Quality 86, 154 - 166 (2013), DOI:10.5073/JABFQ.2013.086.021 1University of Kiel, Institute of Crop Science and Plant Breeding – Grass and Forage Science / Organic Agriculture, Kiel, Germany 2Institute of Ecological Chemistry, Plant Analysis and Stored Product Protection; Julius Kühn-Institute, Quedlinburg, Germany Different approaches to evaluate tannin content and structure of selected plant extracts – review and new aspects M. Kardel1*, F. Taube1, H. Schulz2, W. Schütze2, M. Gierus1 (Received July 2, 2013) Summary extracts, tannins, salts and derivates was imported to the EU in the year 2011, mainly from Argentina, with a trade value of nearly Tannins occur in many field herbs and legumes, providing an im- 63.5 mio. US Dollar (UN-COMTRADE, 2013). These extracts are pro- mense variability in structure and molecular weight. This leads to duced industrially on a large scale. They could therefore maintain complications when measuring tannin content; comparability of a relatively constant quality and thus provide an advantage in rumi- different methods is problematic. The present investigations aimed at nant feeding. characterizing four different tannin extracts: quebracho (Schinopsis Tannins from different sources can react very diverse regarding lorentzii), mimosa (Acacia mearnsii), tara (Caesalpinia spinosa), protein affinity (MCNABB et al., 1998; BUENO et al., 2008). Diffe- and gambier (Uncaria gambir) and impact of storage conditions. rences in tannin structure can occur even between similar plant Using photometrical methods as well as HPLC-ESI-MS, fundamen- species (OSBORNE and MCNEILL, 2001; HATTAS et al., 2011) and a tal differences could be determined. -

Effect of Practical Timing of Dosage on Theophylline Blood Levels In
Arch Dis Child: first published as 10.1136/adc.53.2.167 on 1 February 1978. Downloaded from Archives of Disease in Childhood, 1978, 53, 167-182 Short reports Effect of practical timing of dosage Methods on theophylline blood levels in For the period of the study, the children did not asthmatic children treated with receive tea, which contains theophylline. choline theophyllinate GroupA. These childrenwere given choline theophyll- The bronchodilator effect of theophylline has been inate in a dosage of as near as possible 8 mg/kg per known for over 50 years, but although it has been dose qds (mean 7 9 mg/kg, range 7-2-8 7 mg/kg), widely used both intravenously and rectally in the starting with half the dose on the first day to try and treatment ofacute asthma, its use as an oral broncho- minimise nausea. The doses were given at 0800 h, dilator has been limited by inadequate knowledge of 1300 h, 1700 h, and 2100 h, times considered suitable the correct dosage of the oral preparations. The for administration outside hospital. On the third day therapeutic level of theophylline is generally con- ofthe study serum levels were measured 2 hours after sidered to be between 10 and 20 ,ug/ml (Turner- each dose and before the 0800 h dose the following Warwick, 1957; Jenne et al., 1972), although sub- morning. optimal bronchodilation can be achieved with levels of 5 ,ug/ml or less (Maselli et al., 1970; Nicholson and Group B. These children were given cholinetheophyll- Chick, 1973). Serious toxicity with levels of less than inate in a dosage of as near as possible 10 mg/kg per copyright. -

Epigallocatechin Gallate: a Review
Veterinarni Medicina, 63, 2018 (10): 443–467 Review Article https://doi.org/10.17221/31/2018-VETMED Epigallocatechin gallate: a review L. Bartosikova*, J. Necas Faculty of Medicine and Dentistry, Palacky University, Olomouc, Czech Republic *Corresponding author: [email protected] ABSTRACT: Epigallocatechin gallate is the major component of the polyphenolic fraction of green tea and is responsible for most of the therapeutic benefits of green tea consumption. A number of preclinical in vivo and in vitro experiments as well as clinical trials have shown a wide range of biological and pharmacological properties of polyphenolic compounds such as anti-oxidative, antimicrobial, anti-allergic, anti-diabetic, anti-inflammatory, anti-cancer, chemoprotective, neuroprotective and immunomodulatory effects. Epigallocatechin gallate controls high blood pressure, decreases blood cholesterol and body fat and decreases the risk of osteoporotic fractures. Further research should be performed to monitor the pharmacological and clinical effects of green tea and to more clearly elucidate its mechanisms of action and the potential for its use in medicine. Keywords: epigallocatechin-3-O-gallate; pharmacokinetics; toxicity; biological activity List of abbreviations ADME = absorption, distribution, metabolism, excretion; AMP = activated protein kinase (AMPK); AUC (0–∞) = area under the concentration-time curve from 0 h to infinity; BAX = apoptosis regulator (pro-apoptotic regulator, known as bcl-2-like protein 4); Bcl-2 = B-cell lymphoma 2; BCL-XL = B-cell -

3-Methylxanthine Production Through Biodegradation of Theobromine By
Zhou et al. BMC Microbiology (2020) 20:269 https://doi.org/10.1186/s12866-020-01951-z RESEARCH ARTICLE Open Access 3-Methylxanthine production through biodegradation of theobromine by Aspergillus sydowii PT-2 Binxing Zhou1*† , Cunqiang Ma1,2,3*†, Chengqin Zheng1, Tao Xia4, Bingsong Ma1 and Xiaohui Liu1 Abstract Background: Methylxanthines, including caffeine, theobromine and theophylline, are natural and synthetic compounds in tea, which could be metabolized by certain kinds of bacteria and fungi. Previous studies confirmed that several microbial isolates from Pu-erh tea could degrade and convert caffeine and theophylline. We speculated that these candidate isolates also could degrade and convert theobromine through N-demethylation and oxidation. In this study, seven tea-derived fungal strains were inoculated into various theobromine agar medias and theobromine liquid mediums to assess their capacity in theobromine utilization. Related metabolites with theobromine degradation were detected by using HPLC in the liquid culture to investigate their potential application in the production of 3-methylxanthine. Results: Based on theobromine utilization capacity, Aspergillus niger PT-1, Aspergillus sydowii PT-2, Aspergillus ustus PT-6 and Aspergillus tamarii PT-7 have demonstrated the potential for theobromine biodegradation. Particularly, A. sydowii PT-2 and A. tamarii PT-7 could degrade theobromine significantly (p < 0.05) in all given liquid mediums. 3,7- Dimethyluric acid, 3-methylxanthine, 7-methylxanthine, 3-methyluric acid, xanthine, and uric acid were detected in A. sydowii PT-2 and A. tamarii PT-7 culture, respectively, which confirmed the existence of N-demethylation and oxidation in theobromine catabolism. 3-Methylxanthine was common and main demethylated metabolite of theobromine in the liquid culture. -

Influence of COMT Genotype Polymorphism on Plasma and Urine
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by University of Minnesota Digital Conservancy Influence of COMT Genotype Polymorphism on Plasma and Urine Green Tea Catechin Levels in Postmenopausal Women A THESIS SUBMITTED TO THE FACULTY OF THE UNIVERSITY OF MINNESOTA BY Alyssa Heather Perry IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE Dr. Mindy Kurzer September 2014 © Alyssa Heather Perry 2014 Abstract Catechins are the major polyphenolic compound in green tea that have been investigated extensively over the past few decades in relation to the treatment of various chronic diseases including diabetes, cardiovascular disease, and cancer. O-methylation is a major Phase II metabolic pathway of green tea catechins (GTCs) via the enzyme catechol-O- methyltransferase (COMT). A single nucleotide polymorphism in the gene coding for COMT leads to individuals with a high-, intermediate-, or low-activity COMT enzyme. An epidemiological case-control study suggests that green tea consumption is associated with reduced risk of breast cancer in women with an intermediate- or low-activity COMT genotype. A cross-sectional analysis discovered that men homozygous for the low- activity COMT genotype showed a reduction in total tea polyphenols in spot urine samples compared to the intermediate- and high-activity genotypes. Several human intervention trials have assessed green tea intake, metabolism, and COMT genotype with conflicting results. The aim of the present study was to determine if the COMT polymorphism would modify the excretion and plasma levels of GTCs in 180 postmenopausal women at high risk for breast cancer consuming a green tea extract supplement containing 1222 mg total catechins daily for 12 months. -

Enabling Direct Preferential Crystallization in a Stable Racemic
Enabling Direct Preferential Crystallization in a Stable Racemic Compound System Lina Harfouche, Clément Brandel, Yohann Cartigny, Joop ter Horst, Gérard Coquerel, Samuel Petit To cite this version: Lina Harfouche, Clément Brandel, Yohann Cartigny, Joop ter Horst, Gérard Coquerel, et al.. Enabling Direct Preferential Crystallization in a Stable Racemic Compound System. Molecular Pharmaceutics, American Chemical Society, 2019, 16 (11), pp.4670-4676. 10.1021/acs.molpharmaceut.9b00805. hal- 02331962 HAL Id: hal-02331962 https://hal-normandie-univ.archives-ouvertes.fr/hal-02331962 Submitted on 24 Oct 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Enabling Direct Preferential Crystallization in a Stable Racemic Compound System Lina C. Harfouche,† Clément Brandel,†* Yohann Cartigny,† Joop H. ter Horst,‡ Gérard Coquerel,† and Samuel Petit† † Universite de Rouen Normandie, UFR des Sciences et Techniques, Laboratoire SMS-EA3233, Place Emile Blondel, 76821, Mont-Saint-Aignan, France ‡EPSRC Centre for Innovative Manufacturing in Continuous Manufacturing and Crystallisation (CMAC), Strathclyde Insti- tute of Pharmacy and Biomedical Sciences (SIPBS), Technology and Innovation Centre, University of Strathclyde, 99 George Street, Glasgow G1 1RD (UK) Supporting Information Placeholder 8 ABSTRACT: The preparative resolution by preferential crystal- separation methods, including chiral chromatography and enzy- 9 lization (PC) of proxyphylline has been achieved despite the matic resolution. -
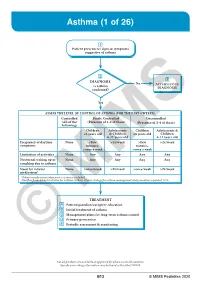
Asthma (1 of 26)
Asthma (1 of 26) 1 Patient presents w/ signs & symptoms suggestive of asthma 2 3 DIAGNOSIS No ALTERNATIVE Is asthma DIAGNOSIS confi rmed? Yes ASSESS THE LEVEL OF CONTROL OF ASTHMA FOR THE PAST 4 WEEKS Controlled Partly Controlled Uncontrolled (All of the (Presence of 1-2 of these) (Presence of 3-4 of these) following) Children Adolescents Children Adolescents & ≤5 years old & Children ≤5 years old Children 6-11 years old 6-11 years old Frequency of daytime None >Few >2x/week >Few >2x/week symptoms minutes, minutes, >once a week >once a week Limitation of activities None Any Any Any Any Nocturnal waking up or None Any Any Any Any coughing due to asthma Need for reliever None >once/week >2x/week >once/week >2x/week medication* *Reliever medications taken prior to exercise excluded. Modified from: Global Initiative for Asthma (GINA). Global strategy for asthma management and prevention: Updated 2020. TREATMENT A Patient/guardian/caregiver education B Initial treatment of asthma C Management plans for long-term asthma control D Primary prevention E © Periodic assessmentMIMS & monitoring Not all products are available or approved for above use in all countries. Specifi c prescribing information may be found in the latest MIMS. B13 © MIMS Pediatrics 2020 Asthma (2 of 26) 1 ASTHMA • A heterogeneous disease w/ chronic infl ammatory disorder of the airways • e most common chronic disease in pediatric age groups that causes signifi cant morbidity • Characterized by history of respiratory symptoms eg wheeze, shortness of breath, chest tightness & cough