Soil Protists in Three Neotropical Rainforests Are Hyperdiverse And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Closest Unicellular Relatives of Animals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 12, 1773–1778, October 15, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01187-9 The Closest Unicellular Relatives of Animals B.F. Lang,1,2 C. O’Kelly,1,3 T. Nerad,4 M.W. Gray,1,5 Results and Discussion and G. Burger1,2,6 1The Canadian Institute for Advanced Research The evolution of the Metazoa from single-celled protists Program in Evolutionary Biology is an issue that has intrigued biologists for more than a 2 De´ partement de Biochimie century. Early morphological and more recent ultra- Universite´ de Montre´ al structural and molecular studies have converged in sup- Succursale Centre-Ville porting the now widely accepted view that animals are Montre´ al, Que´ bec H3C 3J7 related to Fungi, choanoflagellates, and ichthyosporean Canada protists. However, controversy persists as to the spe- 3 Bigelow Laboratory for Ocean Sciences cific evolutionary relationships among these major P.O. Box 475 groups. This uncertainty is reflected in the plethora of 180 McKown Point Road published molecular phylogenies that propose virtually West Boothbay Harbor, Maine 04575 all of the possible alternative tree topologies involving 4 American Type Culture Collection Choanoflagellata, Fungi, Ichthyosporea, and Metazoa. 10801 University Boulevard For example, a monophyletic MetazoaϩChoanoflagel- Manassas, Virginia 20110 lata group has been suggested on the basis of small 5 Department of Biochemistry subunit (SSU) rDNA sequences [1, 6, 7]. Other studies and Molecular Biology using the same sequences have allied Choanoflagellata Dalhousie University with the Fungi [8], placed Choanoflagellata prior to the Halifax, Nova Scotia B3H 4H7 divergence of animals and Fungi [9], or even placed Canada them prior to the divergence of green algae and land plants [10]. -

Ptolemeba N. Gen., a Novel Genus of Hartmannellid Amoebae (Tubulinea, Amoebozoa); with an Emphasis on the Taxonomy of Saccamoeba
The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists Journal of Eukaryotic Microbiology ISSN 1066-5234 ORIGINAL ARTICLE Ptolemeba n. gen., a Novel Genus of Hartmannellid Amoebae (Tubulinea, Amoebozoa); with an Emphasis on the Taxonomy of Saccamoeba Pamela M. Watsona, Stephanie C. Sorrella & Matthew W. Browna,b a Department of Biological Sciences, Mississippi State University, Mississippi State, Mississippi, 39762 b Institute for Genomics, Biocomputing & Biotechnology, Mississippi State University, Mississippi State, Mississippi, 39762 Keywords ABSTRACT 18S rRNA; amoeba; amoeboid; Cashia; cristae; freshwater amoebae; Hartmannella; Hartmannellid amoebae are an unnatural assemblage of amoeboid organisms mitochondrial morphology; SSU rDNA; SSU that are morphologically difficult to discern from one another. In molecular phy- rRNA; terrestrial amoebae; tubulinid. logenetic trees of the nuclear-encoded small subunit rDNA, they occupy at least five lineages within Tubulinea, a well-supported clade in Amoebozoa. The Correspondence polyphyletic nature of the hartmannellids has led to many taxonomic problems, M.W. Brown, Department of Biological in particular paraphyletic genera. Recent taxonomic revisions have alleviated Sciences, Mississippi State University, some of the problems. However, the genus Saccamoeba is paraphyletic and is Mississippi State, MS 39762, USA still in need of revision as it currently occupies two distinct lineages. Here, we Telephone number: +1 662-325-2406; report a new clade on the tree of Tubulinea, which we infer represents a novel FAX number: +1 662-325-7939; genus that we name Ptolemeba n. gen. This genus subsumes a clade of hart- e-mail: [email protected] mannellid amoebae that were previously considered in the genus Saccamoeba, but whose mitochondrial morphology is distinct from Saccamoeba. -

Protistology Mitochondrial Genomes of Amoebozoa
Protistology 13 (4), 179–191 (2019) Protistology Mitochondrial genomes of Amoebozoa Natalya Bondarenko1, Alexey Smirnov1, Elena Nassonova1,2, Anna Glotova1,2 and Anna Maria Fiore-Donno3 1 Department of Invertebrate Zoology, Faculty of Biology, Saint Petersburg State University, 199034 Saint Petersburg, Russia 2 Laboratory of Cytology of Unicellular Organisms, Institute of Cytology RAS, 194064 Saint Petersburg, Russia 3 University of Cologne, Institute of Zoology, Terrestrial Ecology, 50674 Cologne, Germany | Submitted November 28, 2019 | Accepted December 10, 2019 | Summary In this mini-review, we summarize the current knowledge on mitochondrial genomes of Amoebozoa. Amoebozoa is a major, early-diverging lineage of eukaryotes, containing at least 2,400 species. At present, 32 mitochondrial genomes belonging to 18 amoebozoan species are publicly available. A dearth of information is particularly obvious for two major amoebozoan clades, Variosea and Tubulinea, with just one mitochondrial genome sequenced for each. The main focus of this review is to summarize features such as mitochondrial gene content, mitochondrial genome size variation, and presence or absence of RNA editing, showing if they are unique or shared among amoebozoan lineages. In addition, we underline the potential of mitochondrial genomes for multigene phylogenetic reconstruction in Amoebozoa, where the relationships among lineages are not fully resolved yet. With the increasing application of next-generation sequencing techniques and reliable protocols, we advocate mitochondrial -

An Intestinal Gregarine of Nothria Conchylega (Polychaeta, Onuphidae)
Journal of Invertebrate Pathology 104 (2010) 172–179 Contents lists available at ScienceDirect Journal of Invertebrate Pathology journal homepage: www.elsevier.com/locate/jip Description of Trichotokara nothriae n. gen. et sp. (Apicomplexa, Lecudinidae) – An intestinal gregarine of Nothria conchylega (Polychaeta, Onuphidae) Sonja Rueckert *, Brian S. Leander Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity, Departments of Botany and Zoology, University of British Columbia, #3529 – 6270 University Blvd., Vancouver, BC, Canada V6T 1Z4 article info abstract Article history: The trophozoites of a novel gregarine apicomplexan, Trichotokara nothriae n. gen. et sp., were isolated and Received 12 November 2009 characterized from the intestines of the onuphid tubeworm Nothria conchylega (Polychaeta), collected at Accepted 11 March 2010 20 m depth from the North-eastern Pacific Coast. The trophozoites were 50–155 lm long with a mid-cell Available online 23 March 2010 indentation that formed two prominent bulges (anterior bulge, 14–48 lm wide; posterior bulge, 15– 55 lm wide). Scanning electron microscopy (SEM) demonstrated that approximately 400 densely packed, Keywords: longitudinal epicytic folds (5 folds/lm) inscribe the surface of the trophozoites, and a prominently elon- Alveolata gated mucron (14–60 lm long and 6–12 lm wide) was covered with hair-like projections (mean length, Apicomplexa 1.97 m; mean width, 0.2 m at the base). Although a septum occurred at the junction between the cell Lecudinidae l l Lecudina proper and the mucron in most trophozoites, light microscopy (LM) demonstrated that the cell proper Parasite extended into the core of the mucron as a thin prolongation. -
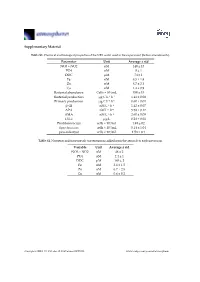
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

A Revised Classification of Naked Lobose Amoebae (Amoebozoa
Protist, Vol. 162, 545–570, October 2011 http://www.elsevier.de/protis Published online date 28 July 2011 PROTIST NEWS A Revised Classification of Naked Lobose Amoebae (Amoebozoa: Lobosa) Introduction together constitute the amoebozoan subphy- lum Lobosa, which never have cilia or flagella, Molecular evidence and an associated reevaluation whereas Variosea (as here revised) together with of morphology have recently considerably revised Mycetozoa and Archamoebea are now grouped our views on relationships among the higher-level as the subphylum Conosa, whose constituent groups of amoebae. First of all, establishing the lineages either have cilia or flagella or have lost phylum Amoebozoa grouped all lobose amoe- them secondarily (Cavalier-Smith 1998, 2009). boid protists, whether naked or testate, aerobic Figure 1 is a schematic tree showing amoebozoan or anaerobic, with the Mycetozoa and Archamoe- relationships deduced from both morphology and bea (Cavalier-Smith 1998), and separated them DNA sequences. from both the heterolobosean amoebae (Page and The first attempt to construct a congruent molec- Blanton 1985), now belonging in the phylum Per- ular and morphological system of Amoebozoa by colozoa - Cavalier-Smith and Nikolaev (2008), and Cavalier-Smith et al. (2004) was limited by the the filose amoebae that belong in other phyla lack of molecular data for many amoeboid taxa, (notably Cercozoa: Bass et al. 2009a; Howe et al. which were therefore classified solely on morpho- 2011). logical evidence. Smirnov et al. (2005) suggested The phylum Amoebozoa consists of naked and another system for naked lobose amoebae only; testate lobose amoebae (e.g. Amoeba, Vannella, this left taxa with no molecular data incertae sedis, Hartmannella, Acanthamoeba, Arcella, Difflugia), which limited its utility. -

Comparative Proteomic Profiling of Newly Acquired, Virulent And
www.nature.com/scientificreports OPEN Comparative proteomic profling of newly acquired, virulent and attenuated Neoparamoeba perurans proteins associated with amoebic gill disease Kerrie Ní Dhufaigh1*, Eugene Dillon2, Natasha Botwright3, Anita Talbot1, Ian O’Connor1, Eugene MacCarthy1 & Orla Slattery4 The causative agent of amoebic gill disease, Neoparamoeba perurans is reported to lose virulence during prolonged in vitro maintenance. In this study, the impact of prolonged culture on N. perurans virulence and its proteome was investigated. Two isolates, attenuated and virulent, had their virulence assessed in an experimental trial using Atlantic salmon smolts and their bacterial community composition was evaluated by 16S rRNA Illumina MiSeq sequencing. Soluble proteins were isolated from three isolates: a newly acquired, virulent and attenuated N. perurans culture. Proteins were analysed using two-dimensional electrophoresis coupled with liquid chromatography tandem mass spectrometry (LC–MS/MS). The challenge trial using naïve smolts confrmed a loss in virulence in the attenuated N. perurans culture. A greater diversity of bacterial communities was found in the microbiome of the virulent isolate in contrast to a reduction in microbial community richness in the attenuated microbiome. A collated proteome database of N. perurans, Amoebozoa and four bacterial genera resulted in 24 proteins diferentially expressed between the three cultures. The present LC–MS/ MS results indicate protein synthesis, oxidative stress and immunomodulation are upregulated in a newly acquired N. perurans culture and future studies may exploit these protein identifcations for therapeutic purposes in infected farmed fsh. Neoparamoeba perurans is an ectoparasitic protozoan responsible for the hyperplastic gill infection of marine cultured fnfsh referred to as amoebic gill disease (AGD)1. -

Predatory Flagellates – the New Recently Discovered Deep Branches of the Eukaryotic Tree and Their Evolutionary and Ecological Significance
Protistology 14 (1), 15–22 (2020) Protistology Predatory flagellates – the new recently discovered deep branches of the eukaryotic tree and their evolutionary and ecological significance Denis V. Tikhonenkov Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, 152742, Russia | Submitted March 20, 2020 | Accepted April 6, 2020 | Summary Predatory protists are poorly studied, although they are often representing important deep-branching evolutionary lineages and new eukaryotic supergroups. This short review/opinion paper is inspired by the recent discoveries of various predatory flagellates, which form sister groups of the giant eukaryotic clusters on phylogenetic trees, and illustrate an ancestral state of one or another supergroup of eukaryotes. Here we discuss their evolutionary and ecological relevance and show that the study of such protists may be essential in addressing previously puzzling evolutionary problems, such as the origin of multicellular animals, the plastid spread trajectory, origins of photosynthesis and parasitism, evolution of mitochondrial genomes. Key words: evolution of eukaryotes, heterotrophic flagellates, mitochondrial genome, origin of animals, photosynthesis, predatory protists, tree of life Predatory flagellates and diversity of eu- of the hidden diversity of protists (Moon-van der karyotes Staay et al., 2000; López-García et al., 2001; Edg- comb et al., 2002; Massana et al., 2004; Richards The well-studied multicellular animals, plants and Bass, 2005; Tarbe et al., 2011; de Vargas et al., and fungi immediately come to mind when we hear 2015). In particular, several prevailing and very abun- the term “eukaryotes”. However, these groups of dant ribogroups such as MALV, MAST, MAOP, organisms represent a minority in the real diversity MAFO (marine alveolates, stramenopiles, opistho- of evolutionary lineages of eukaryotes. -

Multiple Roots of Fruiting Body Formation in Amoebozoa
GBE Multiple Roots of Fruiting Body Formation in Amoebozoa Falk Hillmann1,*, Gillian Forbes2, Silvia Novohradska1, Iuliia Ferling1,KonstantinRiege3,MarcoGroth4, Martin Westermann5,ManjaMarz3, Thomas Spaller6, Thomas Winckler6, Pauline Schaap2,and Gernot Glo¨ ckner7,* 1Junior Research Group Evolution of Microbial Interaction, Leibniz Institute for Natural Product Research and Infection Biology – Hans Kno¨ ll Institute (HKI), Jena, Germany 2Division of Cell and Developmental Biology, School of Life Sciences, University of Dundee, United Kingdom 3Bioinformatics/High Throughput Analysis, Friedrich Schiller University Jena, Germany 4CF DNA-Sequencing, Leibniz Institute on Aging Research, Jena, Germany 5Electron Microscopy Center, Jena University Hospital, Germany 6Pharmaceutical Biology, Institute of Pharmacy, Friedrich Schiller University Jena, Germany 7Institute of Biochemistry I, Medical Faculty, University of Cologne, Germany *Corresponding authors: E-mails: [email protected]; [email protected]. Accepted: January 11, 2018 Data deposition: The genome sequence and gene predictions of Protostelium aurantium and Protostelium mycophagum were deposited in GenBank under the Accession Numbers MDYQ00000000 and MZNV00000000, respectively. The mitochondrial genome of P. mycophagum was deposited under the Accession number KY75056 and that of P. aurantium under the Accession number KY75057. The RNAseq reads can be found in Bioproject Accession PRJNA338377. All sequence and annotation data are also available directly from the authors. The P. aurantium strain is deposited in the Jena Microbial Resource Collection (JMRC) under accession number SF0012540. Abstract Establishment of multicellularity represents a major transition in eukaryote evolution. A subgroup of Amoebozoa, the dictyos- teliids, has evolved a relatively simple aggregative multicellular stage resulting in a fruiting body supported by a stalk. Protosteloid amoeba, which are scattered throughout the amoebozoan tree, differ by producing only one or few single stalked spores. -

The Revised Classification of Eukaryotes
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/231610049 The Revised Classification of Eukaryotes Article in Journal of Eukaryotic Microbiology · September 2012 DOI: 10.1111/j.1550-7408.2012.00644.x · Source: PubMed CITATIONS READS 961 2,825 25 authors, including: Sina M Adl Alastair Simpson University of Saskatchewan Dalhousie University 118 PUBLICATIONS 8,522 CITATIONS 264 PUBLICATIONS 10,739 CITATIONS SEE PROFILE SEE PROFILE Christopher E Lane David Bass University of Rhode Island Natural History Museum, London 82 PUBLICATIONS 6,233 CITATIONS 464 PUBLICATIONS 7,765 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Biodiversity and ecology of soil taste amoeba View project Predator control of diversity View project All content following this page was uploaded by Smirnov Alexey on 25 October 2017. The user has requested enhancement of the downloaded file. The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists J. Eukaryot. Microbiol., 59(5), 2012 pp. 429–493 © 2012 The Author(s) Journal of Eukaryotic Microbiology © 2012 International Society of Protistologists DOI: 10.1111/j.1550-7408.2012.00644.x The Revised Classification of Eukaryotes SINA M. ADL,a,b ALASTAIR G. B. SIMPSON,b CHRISTOPHER E. LANE,c JULIUS LUKESˇ,d DAVID BASS,e SAMUEL S. BOWSER,f MATTHEW W. BROWN,g FABIEN BURKI,h MICAH DUNTHORN,i VLADIMIR HAMPL,j AARON HEISS,b MONA HOPPENRATH,k ENRIQUE LARA,l LINE LE GALL,m DENIS H. LYNN,n,1 HILARY MCMANUS,o EDWARD A. D. -

S41467-021-25308-W.Pdf
ARTICLE https://doi.org/10.1038/s41467-021-25308-w OPEN Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota ✉ ✉ Luis Javier Galindo 1 , Purificación López-García 1, Guifré Torruella1, Sergey Karpov2,3 & David Moreira 1 Compared to multicellular fungi and unicellular yeasts, unicellular fungi with free-living fla- gellated stages (zoospores) remain poorly known and their phylogenetic position is often 1234567890():,; unresolved. Recently, rRNA gene phylogenetic analyses of two atypical parasitic fungi with amoeboid zoospores and long kinetosomes, the sanchytrids Amoeboradix gromovi and San- chytrium tribonematis, showed that they formed a monophyletic group without close affinity with known fungal clades. Here, we sequence single-cell genomes for both species to assess their phylogenetic position and evolution. Phylogenomic analyses using different protein datasets and a comprehensive taxon sampling result in an almost fully-resolved fungal tree, with Chytridiomycota as sister to all other fungi, and sanchytrids forming a well-supported, fast-evolving clade sister to Blastocladiomycota. Comparative genomic analyses across fungi and their allies (Holomycota) reveal an atypically reduced metabolic repertoire for sanchy- trids. We infer three main independent flagellum losses from the distribution of over 60 flagellum-specific proteins across Holomycota. Based on sanchytrids’ phylogenetic position and unique traits, we propose the designation of a novel phylum, Sanchytriomycota. In addition, our results indicate that most of the hyphal morphogenesis gene repertoire of multicellular fungi had already evolved in early holomycotan lineages. 1 Ecologie Systématique Evolution, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France. 2 Zoological Institute, Russian Academy of Sciences, St. ✉ Petersburg, Russia. 3 St. -

Diptera: Sciaroidea Incertae Sedis), with Description of Three New Species from India and Vietnam
ACTA ENTOMOLOGICA MUSEI NATIONALIS PRAGAE Published 15.xii.2014 Volume 54(2), pp. 729–739 ISSN 0374-1036 http://zoobank.org/urn:lsid:zoobank.org:pub:8FF046B2-6267-454D-8A28-9AE80CC0F23C Notes on Nepaletricha (Diptera: Sciaroidea incertae sedis), with description of three new species from India and Vietnam Heikki HIPPA1) & Jan ŠEVýÍK2) 1) Gribbylunds allé 2, SE-183 65 Täby, Sweden; e-mail: [email protected] 2) Department of Biology and Ecology, University of Ostrava, Chittussiho 10, CZ-71000 Ostrava, Czech Republic; e-mail: [email protected] Abstract. The following new species are described: Nepaletricha dembickyi sp. nov. (India), N. lobosa sp. nov. (Vietnam) and N. sigma sp. nov. (India). The sys- tematic position of the genus is brieÀ y discussed. A key to the six known species of Nepaletricha is given. Key words. Insecta, Diptera, Sciaroidea, Antefungivoridae, Pleciomimidae, Rangomaramidae, Heterotricha group, Nepaletricha, fungus gnats, taxonomy, key, Oriental Region Introduction The genus Nepaletricha was established by CHANDLER (2002) for a single Oriental species Nepaletricha mystica Chandler, 2002. Subsequently, HIPPA et al. (2009) re-characterized the genus and added two new species from northern Thailand and Vietnam, respectively. The genus was considered as Sciaroidea incertae sedis by CHANDLER (2002) and HIPPA & VILKAMAA (2005, 2006). AMORIM & RINDAL (2007) placed Nepaletricha in the family Rangomaramidae, together with many other earlier unplaced genera of Sciaroidea. Concerning Nepaletricha, HIPPA et al. (2009) followed this placement. The classi¿ cation of AMORIM & RINDAL (2007) was strongly criticized by JASCHHOF (2011) who did not accept the new concept of Rangomara- midae and suggested to continue using Sciaroidea incertae sedis for a number of problematic genera of Sciaroidea.