Microbial Eukaryotes in Oil Sands Environments: Heterotrophs in the Spotlight
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protozoologica Special Issue: Protists in Soil Processes
Acta Protozool. (2012) 51: 201–208 http://www.eko.uj.edu.pl/ap ActA doi:10.4467/16890027AP.12.016.0762 Protozoologica Special issue: Protists in Soil Processes Review paper Ecology of Soil Eumycetozoans Steven L. STEPHENSON1 and Alan FEEST2 1Department of Biological Sciences, University of Arkansas, Fayetteville, Arkansas, USA; 2Institute of Advanced Studies, University of Bristol and Ecosulis ltd., Newton St Loe, Bath, United Kingdom Abstract. Eumycetozoans, commonly referred to as slime moulds, are common to abundant organisms in soils. Three groups of slime moulds (myxogastrids, dictyostelids and protostelids) are recognized, and the first two of these are among the most important bacterivores in the soil microhabitat. The purpose of this paper is first to provide a brief description of all three groups and then to review what is known about their distribution and ecology in soils. Key words: Amoebae, bacterivores, dictyostelids, myxogastrids, protostelids. INTRODUCTION that they are amoebozoans and not fungi (Bapteste et al. 2002, Yoon et al. 2008, Baudalf 2008). Three groups of slime moulds (myxogastrids, dic- One of the idiosyncratic branches of the eukary- tyostelids and protostelids) are recognized (Olive 1970, otic tree of life consists of an assemblage of amoe- 1975). Members of the three groups exhibit consider- boid protists referred to as the supergroup Amoebozoa able diversity in the type of aerial spore-bearing struc- (Fiore-Donno et al. 2010). The most diverse members tures produced, which can range from exceedingly of the Amoebozoa are the eumycetozoans, common- small examples (most protostelids) with only a single ly referred to as slime moulds. Since their discovery, spore to the very largest examples (certain myxogas- slime moulds have been variously classified as plants, trids) that contain many millions of spores. -

Comparative Genomics of the Social Amoebae Dictyostelium Discoideum
Sucgang et al. Genome Biology 2011, 12:R20 http://genomebiology.com/2011/12/2/R20 RESEARCH Open Access Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum Richard Sucgang1†, Alan Kuo2†, Xiangjun Tian3†, William Salerno1†, Anup Parikh4, Christa L Feasley5, Eileen Dalin2, Hank Tu2, Eryong Huang4, Kerrie Barry2, Erika Lindquist2, Harris Shapiro2, David Bruce2, Jeremy Schmutz2, Asaf Salamov2, Petra Fey6, Pascale Gaudet6, Christophe Anjard7, M Madan Babu8, Siddhartha Basu6, Yulia Bushmanova6, Hanke van der Wel5, Mariko Katoh-Kurasawa4, Christopher Dinh1, Pedro M Coutinho9, Tamao Saito10, Marek Elias11, Pauline Schaap12, Robert R Kay8, Bernard Henrissat9, Ludwig Eichinger13, Francisco Rivero14, Nicholas H Putnam3, Christopher M West5, William F Loomis7, Rex L Chisholm6, Gad Shaulsky3,4, Joan E Strassmann3, David C Queller3, Adam Kuspa1,3,4* and Igor V Grigoriev2 Abstract Background: The social amoebae (Dictyostelia) are a diverse group of Amoebozoa that achieve multicellularity by aggregation and undergo morphogenesis into fruiting bodies with terminally differentiated spores and stalk cells. There are four groups of dictyostelids, with the most derived being a group that contains the model species Dictyostelium discoideum. Results: We have produced a draft genome sequence of another group dictyostelid, Dictyostelium purpureum, and compare it to the D. discoideum genome. The assembly (8.41 × coverage) comprises 799 scaffolds totaling 33.0 Mb, comparable to the D. discoideum genome size. Sequence comparisons suggest that these two dictyostelids shared a common ancestor approximately 400 million years ago. In spite of this divergence, most orthologs reside in small clusters of conserved synteny. Comparative analyses revealed a core set of orthologous genes that illuminate dictyostelid physiology, as well as differences in gene family content. -

The Closest Unicellular Relatives of Animals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 12, 1773–1778, October 15, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01187-9 The Closest Unicellular Relatives of Animals B.F. Lang,1,2 C. O’Kelly,1,3 T. Nerad,4 M.W. Gray,1,5 Results and Discussion and G. Burger1,2,6 1The Canadian Institute for Advanced Research The evolution of the Metazoa from single-celled protists Program in Evolutionary Biology is an issue that has intrigued biologists for more than a 2 De´ partement de Biochimie century. Early morphological and more recent ultra- Universite´ de Montre´ al structural and molecular studies have converged in sup- Succursale Centre-Ville porting the now widely accepted view that animals are Montre´ al, Que´ bec H3C 3J7 related to Fungi, choanoflagellates, and ichthyosporean Canada protists. However, controversy persists as to the spe- 3 Bigelow Laboratory for Ocean Sciences cific evolutionary relationships among these major P.O. Box 475 groups. This uncertainty is reflected in the plethora of 180 McKown Point Road published molecular phylogenies that propose virtually West Boothbay Harbor, Maine 04575 all of the possible alternative tree topologies involving 4 American Type Culture Collection Choanoflagellata, Fungi, Ichthyosporea, and Metazoa. 10801 University Boulevard For example, a monophyletic MetazoaϩChoanoflagel- Manassas, Virginia 20110 lata group has been suggested on the basis of small 5 Department of Biochemistry subunit (SSU) rDNA sequences [1, 6, 7]. Other studies and Molecular Biology using the same sequences have allied Choanoflagellata Dalhousie University with the Fungi [8], placed Choanoflagellata prior to the Halifax, Nova Scotia B3H 4H7 divergence of animals and Fungi [9], or even placed Canada them prior to the divergence of green algae and land plants [10]. -

Future Supply of Oil and Gas from the Gulf of Mexico
Future Supply of Oil and Gas From the Gulf of Mexico U.S. GEOLOGICAL SUltyEY PROFESSIONAL PAPER 1294 Future Supply of Oil and Gas From the Gulf of Mexico By E. D. Attanasi and]. L. Haynes U.S. GEOLOGICAL SURVEY PROFESSIONAL PAPER 1294 An engineering-economic costing algorithm combined with a discovery process model to forecast long-run incremental costs of undiscovered oil and gas UNITED STATES GOVERNMENT PRINTING OFFICE, WASHINGTON : 1983 UNITED STATES DEPARTMENT OF THE INTERIOR JAMES G. WATT, Secretary GEOLOGICAL SURVEY Dallas L. Peck, Director Library of Congress Cataloging in Publication Data Attanasi, E. D. Future supply of oil and gas from the Gulf of Mexico. (U.S. Geological Survey professional paper ; 1294) Bibliography: p. 1. Petroleum in submerged lands Mexico, Gulf of. 2. Gas, Natural, in submerged lands Mexico, Gulf of. I. Haynes, J. (John), 1954- . II. Title. III. Series: Geological Survey professional paper ; 1294. TN872.A5A87 1983 553.2'8'0916364 83-600030 ____ ____________ For sale by the Superintendent of Documents, U.S. Government Printing Office Washington, D.C. 20402 CONTENTS Page Abstract 1 Introduction 1 Engineering-economic model 3 Methodology 3 Engineering data and assumptions 5 Field classification 5 Field design 6 Production schedules of oil and nonassociated gas wells 7 Economic assumptions and variables 8 Field development costs 8 Production costs and production related taxes 9 Assumptions for after-tax net present value calculations 10 Exploration costs 10 Industry behavior and market conditions 10 Forecasting future discoveries 11 Discovery process model 11 Estimated marginal cost functions for undiscovered recoverable oil and gas resources in the Gulf of Mexico 12 Conclusions and implications 16 References cited 16 Appendix A 17 Appendix B 20 ILLUSTRATIONS FIGURE 1. -
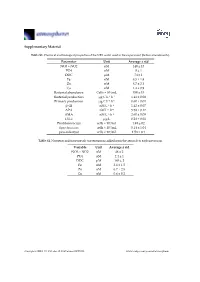
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Predatory Flagellates – the New Recently Discovered Deep Branches of the Eukaryotic Tree and Their Evolutionary and Ecological Significance
Protistology 14 (1), 15–22 (2020) Protistology Predatory flagellates – the new recently discovered deep branches of the eukaryotic tree and their evolutionary and ecological significance Denis V. Tikhonenkov Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, 152742, Russia | Submitted March 20, 2020 | Accepted April 6, 2020 | Summary Predatory protists are poorly studied, although they are often representing important deep-branching evolutionary lineages and new eukaryotic supergroups. This short review/opinion paper is inspired by the recent discoveries of various predatory flagellates, which form sister groups of the giant eukaryotic clusters on phylogenetic trees, and illustrate an ancestral state of one or another supergroup of eukaryotes. Here we discuss their evolutionary and ecological relevance and show that the study of such protists may be essential in addressing previously puzzling evolutionary problems, such as the origin of multicellular animals, the plastid spread trajectory, origins of photosynthesis and parasitism, evolution of mitochondrial genomes. Key words: evolution of eukaryotes, heterotrophic flagellates, mitochondrial genome, origin of animals, photosynthesis, predatory protists, tree of life Predatory flagellates and diversity of eu- of the hidden diversity of protists (Moon-van der karyotes Staay et al., 2000; López-García et al., 2001; Edg- comb et al., 2002; Massana et al., 2004; Richards The well-studied multicellular animals, plants and Bass, 2005; Tarbe et al., 2011; de Vargas et al., and fungi immediately come to mind when we hear 2015). In particular, several prevailing and very abun- the term “eukaryotes”. However, these groups of dant ribogroups such as MALV, MAST, MAOP, organisms represent a minority in the real diversity MAFO (marine alveolates, stramenopiles, opistho- of evolutionary lineages of eukaryotes. -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

The Economic Impacts of the Gulf of Mexico Oil and Natural Gas Industry
The Economic Impacts of the Gulf of Mexico Oil and Natural Gas Industry Prepared For Prepared By Executive Summary Introduction Despite the current difficulties facing the global economy as a whole and the oil and natural gas industry specifically, the Gulf of Mexico oil and natural gas industry will likely continue to be a major source of energy production, employment, gross domestic product, and government revenues for the United States. Several proposals have been advanced recently which would have a major impact on the industry’s activity levels, and the economic activity supported by the Gulf of Mexico offshore oil and natural gas industry. The proposals vary widely, but for the purpose of this report three scenarios were developed, a scenario based on a continuation of current policies and regulations, a scenario examining the potential impacts of a ban on new offshore leases, and a scenario examining the potential impacts of a ban on new drilling permits approvals in the Gulf of Mexico. Energy and Industrial Advisory Partners (EIAP) was commissioned by the National Ocean Industry Association (NOIA) to develop a report forecasting activity levels, spending, oil and natural gas production, supported employment, GDP, and Government Revenues in these scenarios. The scenarios developed in this report are based solely upon government and other publicly available data and EIAP’s own expertise and analysis. The study also included profiles of NOIA members to demonstrate the diverse group of companies which make up the offshore Gulf of Mexico oil and natural gas industry as well as a list of over 2,400 suppliers to the industry representing all 50 states. -

GIULIA MAGRI RIBEIRO Sequenciamento E
GIULIA MAGRI RIBEIRO Sequenciamento e anota¸c˜ao do transcriptoma da ameba tecada Arcella intermedia: Descri¸c˜ao de vias e descobertas de genes. Transcriptome sequencing and annotation of the testate amoeba Arcella intermedia: Pathway description and gene discovery. S˜aoPaulo 2018 GIULIA MAGRI RIBEIRO Sequenciamento e anota¸c˜ao do transcriptoma da ameba tecada Arcella intermedia: Descri¸c˜ao de vias e descobertas de genes. Transcriptome sequencing and annotation of the testate amoeba Arcella intermedia: Pathway description and gene discovery. Disserta¸c˜aoapresentada ao Instituto de Biociˆencias da Universidade de S˜ao Paulo para obten¸c˜aodo t´ıtulo de Mestre em Zoologia pelo Programa de P´os-gradua¸c˜ao em Zoologia. Vers˜ao corrigida contendo as altera¸c˜oes solicitadas pela comiss˜aojulgadora em 30 de Outubro de 2018. A vers˜aooriginal encontra-se no Instituto de Biociˆencias da USP e na Biblioteca Digital de Teses e Disserta¸c˜oes da USP. Supervisor: Prof. Dr. Daniel J.G. Lahr S˜aoPaulo 2018 Ficha catalográfica elaborada pelo Serviço de Biblioteca do Instituto de Biociências da USP, com os dados fornecidos pelo autor no formulário: http://www.ib.usp.br/biblioteca/ficha-catalografica/ficha.php Ribeiro, Giulia Magri Sequenciamento e anotação do transcriptoma da ameba tecada Arcella intermedia: Descrição de vias e descobertas de genes. / Giulia Magri Ribeiro; orientador Daniel José Galafasse Lahr. -- São Paulo, 2018. 118 f. Dissertação (Mestrado) - Instituto de Biociências da Universidade de São Paulo, Departamento de Zoologia. 1. Teca ameba. 2. Transcriptoma. 3. Transferências laterais de genes. 4. Metabolismo anaeróbio. I. Lahr, Daniel José Galafasse, orient. -

S41467-021-25308-W.Pdf
ARTICLE https://doi.org/10.1038/s41467-021-25308-w OPEN Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota ✉ ✉ Luis Javier Galindo 1 , Purificación López-García 1, Guifré Torruella1, Sergey Karpov2,3 & David Moreira 1 Compared to multicellular fungi and unicellular yeasts, unicellular fungi with free-living fla- gellated stages (zoospores) remain poorly known and their phylogenetic position is often 1234567890():,; unresolved. Recently, rRNA gene phylogenetic analyses of two atypical parasitic fungi with amoeboid zoospores and long kinetosomes, the sanchytrids Amoeboradix gromovi and San- chytrium tribonematis, showed that they formed a monophyletic group without close affinity with known fungal clades. Here, we sequence single-cell genomes for both species to assess their phylogenetic position and evolution. Phylogenomic analyses using different protein datasets and a comprehensive taxon sampling result in an almost fully-resolved fungal tree, with Chytridiomycota as sister to all other fungi, and sanchytrids forming a well-supported, fast-evolving clade sister to Blastocladiomycota. Comparative genomic analyses across fungi and their allies (Holomycota) reveal an atypically reduced metabolic repertoire for sanchy- trids. We infer three main independent flagellum losses from the distribution of over 60 flagellum-specific proteins across Holomycota. Based on sanchytrids’ phylogenetic position and unique traits, we propose the designation of a novel phylum, Sanchytriomycota. In addition, our results indicate that most of the hyphal morphogenesis gene repertoire of multicellular fungi had already evolved in early holomycotan lineages. 1 Ecologie Systématique Evolution, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France. 2 Zoological Institute, Russian Academy of Sciences, St. ✉ Petersburg, Russia. 3 St. -

Systematic Index
Systematic Index The systematic index contains the scientific names of all taxa mentioned in the book e.g., Anisonema sp., Anopheles and the vernacular names of protists, for example, tintinnids. The index is two-sided, that is, species ap - pear both with the genus-group name first e.g., Acineria incurvata and with the species-group name first ( incurvata , Acineria ). Species and genera, valid and invalid, are in italics print. The scientific name of a subgenus, when used with a binomen or trinomen, must be interpolated in parentheses between the genus-group name and the species- group name according to the International Code of Zoological Nomenclature. In the following index, these paren - theses are omitted to simplify electronic sorting. Thus, the name Apocolpodidium (Apocolpodidium) etoschense is list - ed as Apocolpodidium Apocolpodidium etoschense . Note that this name is also listed under “ Apocolpodidium etoschense , Apocolpodidium ” and “ etoschense , Apocolpodidium Apocolpodidium ”. Suprageneric taxa, communities, and vernacular names are represented in normal type. A boldface page number indicates the beginning of a detailed description, review, or discussion of a taxon. f or ff means include the following one or two page(s), respectively. A Actinobolina vorax 84 Aegyriana paroliva 191 abberans , Euplotes 193 Actinobolina wenrichii 84 aerophila , Centropyxis 87, 191 abberans , Frontonia 193 Actinobolonidae 216 f aerophila sphagnicola , Centropyxis 87 abbrevescens , Deviata 140, 200, 212 Actinophrys sol 84 aerophila sylvatica -

A Seismic-Reflection Investigation of Gas Hydrates and Sea-Floor Features
A seismic-reflection investigation of gas hydrates and sea-floor features of the upper continental slope of the Garden Banks and Green Canyon regions, northern Gulf of Mexico: Report for cruise G1-99-GM (99002) by Alan Cooper1, David Twichell2, Patrick Hart1 Open-File Report 99-570 1999 This report is preliminary and has not been reviewed for conformity with U.S. Geological Survey editorial standards or with the North American Stratigraphic Code. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. U.S. DEPARTMENT OF THE INTERIOR U.S. GEOLOGICAL SURVEY 1Menlo Park, California 2Woods Hole, Massachusetts Introduction During April 1999, the U.S. Geological Survey (USGS) conducted a 13-day cruise in the Garden Banks and Green Canyon regions of the Gulf of Mexico (Figures 1 and 2). The R/V Gyre, owned by Texas A&M University, was chartered for the cruise. The general objectives were (1) to acquire very high resolution seismic-reflection data and side-scan sonar images of the upper and middle continental slope (200-1200-m water depths), (2) to study the acoustic character and features of the sea floor for evidence of sea-floor hazards, and (3) to look for evidence of subsurface gas hydrates and their effects. The Gulf of Mexico is well known for hydrocarbon resources, with emphasis now on frontier deep-water areas. For water depths greater than about 250 m, the pressure-termperature conditions are correct for the development of shallow-subsurface gas hydrate formation (Anderson et al., 1992).