TCR Affinity Biases Th Cell Differentiation by Regulating CD25, Eef1e1, and Gbp2 Dmitri I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -
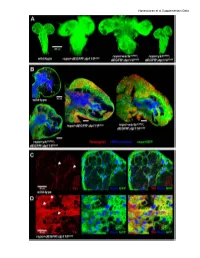
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

Comparative Transcriptome Analysis Reveals Different Host Cell Responses to Acute and Persistent Foot-And-Mouth Disease Virus Infection
Virologica Sinica www.virosin.org https://doi.org/10.1007/s12250-019-00155-8 www.springer.com/12250 (0123456789().,-volV)(0123456789().,-volV) RESEARCH ARTICLE Comparative Transcriptome Analysis Reveals Different Host Cell Responses to Acute and Persistent Foot-and-Mouth Disease Virus Infection 1 1 1 1 1 1 1 Jiadai Li • Lingling Han • Yao Hao • Yuncong Yuan • Mingzhen Wang • Xiu Xin • Hailong Wang • 2 1,3 1,3 Fang Yu • Congyi Zheng • Chao Shen Received: 15 March 2019 / Accepted: 3 July 2019 Ó Wuhan Institute of Virology, CAS 2019 Abstract Foot-and-mouth disease virus (FMDV) rapidly causes cytopathic effects in susceptible cells. Incomplete viral clearance during the acute infection leads to persistent infection. The relationship between host gene expression and the persistent infection remains unclear. In this study, we analyzed the transcriptome profiles of BHK-21 cells acutely and persistently infected with FMDV to identify differences in gene expression. GO and KEGG enrichment analyses showed that the 8,378 differentially expressed genes were significantly enriched in categories including metabolism, biosynthesis, ribosome function, and endocytosis. In persistently infected BHK-21 cells, ribosome- and translation-related genes were significantly down-regulated. There were more differentially expressed immune-related genes during persistent infection than during acute infection. Two hundred and seventy-four genes were differentially expressed in both acutely and persistently infected BHK-21 cells. Among these genes, heat shock protein family B member 1 (Hspb1) knockdown significantly inhibited FMDV replication. Our research provides a basis for further research to understand the mechanisms of persistent FMDV infection including the genes involved in FMDV replication. -

Data-Driven Analysis of Age, Sex, and Tissue Effects on Gene Expression Variability in Alzheimer’S Disease Lavida R.K
bioRxiv preprint doi: https://doi.org/10.1101/498527; this version posted December 17, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Data-Driven Analysis of Age, Sex, and Tissue Effects on Gene Expression Variability in Alzheimer’s Disease Lavida R.K. Brooks 1, George I. Mias,2;∗ 1 Institute for Quantitative Health Science and Engineering, Michigan State University, Microbiology and Molecular Genetics, East Lansing, 48824, USA 2 Institute for Quantitative Health Science and Engineering, Michigan State University, Biochemistry and Molecular Biology, East Lansing, 48824, USA Correspondence*: George I. Mias [email protected] 2 ABSTRACT 3 Alzheimer’s disease (AD) has been categorized by the Centers for Disease Control and 4 Prevention (CDC) as the 6th leading cause of death in the United States. AD is a significant 5 health-care burden because of its increased occurrence (specifically in the elderly population) 6 and the lack of effective treatments and preventive methods. With an increase in life expectancy, 7 the CDC expects AD cases to rise to 15 million by 2060. Aging has been previously associated 8 with susceptibility to AD, and there are ongoing efforts to effectively differentiate between normal 9 and AD age-related brain degeneration and memory loss. AD targets neuronal function and can 10 cause neuronal loss due to the buildup of amyloid-beta plaques and intracellular neurofibrillary 11 tangles. 12 Our study aims to identify temporal changes within gene expression profiles of healthy controls 13 and AD subjects. -

Global Analysis of Trna and Translation Factor Expression Reveals a Dynamic Landscape of Translational Regulation in Human Cancers
ARTICLE https://doi.org/10.1038/s42003-018-0239-8 OPEN Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers Zhao Zhang1, Youqiong Ye1, Jing Gong1, Hang Ruan1, Chun-Jie Liu2, Yu Xiang1, Chunyan Cai3, An-Yuan Guo2, 1234567890():,; Jiqiang Ling4, Lixia Diao5, John N. Weinstein5 & Leng Han 1,6 The protein translational system, including transfer RNAs (tRNAs) and several categories of enzymes, plays a key role in regulating cell proliferation. Translation dysregulation also contributes to cancer development, though relatively little is known about the changes that occur to the translational system in cancer. Here, we present global analyses of tRNAs and three categories of enzymes involved in translational regulation in ~10,000 cancer patients across 31 cancer types from The Cancer Genome Atlas. By analyzing the expression levels of tRNAs at the gene, codon, and amino acid levels, we identified unequal alterations in tRNA expression, likely due to the uneven distribution of tRNAs decoding different codons. We find that overexpression of tRNAs recognizing codons with a low observed-over-expected ratio may overcome the translational bottleneck in tumorigenesis. We further observed overall overexpression and amplification of tRNA modification enzymes, aminoacyl-tRNA synthe- tases, and translation factors, which may play synergistic roles with overexpression of tRNAs to activate the translational systems across multiple cancer types. 1 Department of Biochemistry and Molecular Biology, McGovern Medical School at The University of Texas Health Science Center at Houston, Houston, TX 77030, USA. 2 Department of Bioinformatics and Systems Biology, Hubei Bioinformatics and Molecular Imaging Key Laboratory, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology Wuhan, 430074 Hubei, People’s Republic of China. -

Rare Disruptive Variants in the DISC1 Interactome and Regulome: Association with Cognitive Ability and Schizophrenia
OPEN Molecular Psychiatry (2018) 23, 1270–1277 www.nature.com/mp ORIGINAL ARTICLE Rare disruptive variants in the DISC1 Interactome and Regulome: association with cognitive ability and schizophrenia S Teng1,2,10, PA Thomson3,4,10, S McCarthy1,10, M Kramer1, S Muller1, J Lihm1, S Morris3, DC Soares3, W Hennah5, S Harris3,4, LM Camargo6, V Malkov7, AM McIntosh8, JK Millar3, DH Blackwood8, KL Evans4, IJ Deary4,9, DJ Porteous3,4 and WR McCombie1 Schizophrenia (SCZ), bipolar disorder (BD) and recurrent major depressive disorder (rMDD) are common psychiatric illnesses. All have been associated with lower cognitive ability, and show evidence of genetic overlap and substantial evidence of pleiotropy with cognitive function and neuroticism. Disrupted in schizophrenia 1 (DISC1) protein directly interacts with a large set of proteins (DISC1 Interactome) that are involved in brain development and signaling. Modulation of DISC1 expression alters the expression of a circumscribed set of genes (DISC1 Regulome) that are also implicated in brain biology and disorder. Here we report targeted sequencing of 59 DISC1 Interactome genes and 154 Regulome genes in 654 psychiatric patients and 889 cognitively-phenotyped control subjects, on whom we previously reported evidence for trait association from complete sequencing of the DISC1 locus. Burden analyses of rare and singleton variants predicted to be damaging were performed for psychiatric disorders, cognitive variables and personality traits. The DISC1 Interactome and Regulome showed differential association across the phenotypes tested. After family-wise error correction across all traits (FWERacross), an increased burden of singleton disruptive variants in the Regulome was associated with SCZ (FWERacross P=0.0339). -

Lkb1 Loss Induces Characteristic Pathway Activation in Human
LKB1 LOSS INDUCES CHARACTERISTIC PATHWAY ACTIVATION IN HUMAN TUMORS AND CONFERS SENSITIVITY TO MEK INHIBITION ASSOCIATED WITH ATTENUATED PI3K-AKT-FOXO3 SIGNALING By JACOB KAUFMAN Dissertation Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Cancer Biology December, 2013 Nashville, Tennessee Approved: Professor William Pao Professor Alissa M. Weaver Professor Ethan Lee Professor Pierre P. Massion Professor David P. Carbone This work is dedicated, with love, to my amazing wife, Elizabeth and To our three wonderful daughters, Claire, Caroline, and Mary ii!! ACKNOWLEDGMENTS This work would not have been possible without the financial support from Vanderbilt’s Medical Scientist Training Program and the Strategic Partnering to Evaluate Cancer Signatures grant NCI U01CA114771. It is a privilege to have had such resources available to fund my training and education. I am deeply grateful for the incredible support of my mentor, David Carbone. Dr. Carbone has shared not only his inexhaustible knowledge of biology and cancer, but also his dedication to improving the lives of others, and his sincere compassion both for patients and colleagues. Working with him has given me a model of what being a physician scientist should be, and I hope to emulate his character and compassion throughout my career. This work could not have been completed without the help of my co-authors, especially Joe Amann and Paul Park, as well as Horton Li, Rajesh Arasada and Yu Shyr. I have also benefited immeasurably from many faculty members with whom I’ve had the honor of working and interacting, especially Yu Shyr, Pierre Massion and David Johnson, and my committee members William Pao, Alissa Weaver, and Ethan Lee. -

Cell Division & Proliferation
ptglab.com 1 ANTIBODIES FOR CELL BIOLOGY CELL DIVISION & PROLIFERATION www.ptglab.com 2 Antibodies For Cell Biology: Cell Division & Proliferation Front & Back Cover: Immunohistochemistry of paraffin-embedded human skin tissue slide using Involucrin Antibody (55328-1-AP) at a dilution of 1:50 (10x objective). ptglab.com 3 WELCOME Foreword Cell division and proliferation are processes essential to life; they are processes that underpin growth and development and tissue maintenance. Cell division is the process by which cells must partition their DNA and cell contents appropriately into new daughter cells. This intricate mechanism enables cell proliferation, which must be tightly regulated in balance with cell loss through apoptosis or necrosis. These two processes can go wrong, leading to pathological proliferation, and in so doing form the basis of neoplastic disease. Throughout this catalog you will find hundreds The processes of cell division and proliferation of Proteintech®* antibodies relevant to cell require a wealth of proteins, as demonstrated division and proliferation. They cover mitosis by the long list of components above; if you and meiosis, DNA replication, transcription cannot identify an antibody for your target and translation, as well as protein expression, protein in these pages, please visit our website folding and trafficking and nutrient sensing at ptglab.com to search our full catalog of over and metabolism. There are also antibodies 10,000 pre-made antibodies. We’re confident against cell cycle control proteins featured; you will find the antibody you’re looking for. for instance you will find a poster dedicated to BRCA signaling, a pathway deregulated in many cancers. -

Inflammatory and Adipogenic Stromal Subpopulations in Visceral Adipose Tissue Of
RESEARCH COMMUNICATION Identification of functionally distinct fibro- inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice Chelsea Hepler1†, Bo Shan1, Qianbin Zhang1, Gervaise H Henry2, Mengle Shao1, Lavanya Vishvanath1, Alexandra L Ghaben1, Angela B Mobley3, Douglas Strand2, Gary C Hon4, Rana K Gupta1* 1Touchstone Diabetes Center, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, United States; 2Department of Urology, University of Texas Southwestern Medical Center, Dallas, United States; 3Department of Immunology, University of Texas Southwestern Medical Center, Dallas, United States; 4Cecil H. and Ida Green Center for Reproductive Biology Sciences, Division of Basic Reproductive Biology Research, Department of Obstetrics and Gynecology, University of Texas Southwestern Medical Center, Dallas, United States Abstract White adipose tissue (WAT) remodeling is dictated by coordinated interactions between adipocytes and resident stromal-vascular cells; however, the functional heterogeneity of adipose stromal cells has remained unresolved. We combined single-cell RNA-sequencing and FACS to identify and isolate functionally distinct subpopulations of PDGFRb+ stromal cells within visceral WAT of adult mice. LY6C- CD9- PDGFRb+ cells represent highly adipogenic visceral adipocyte precursor cells (‘APCs’), whereas LY6C+ PDGFRb+ cells represent fibro-inflammatory progenitors (‘FIPs’). FIPs lack adipogenic capacity, display pro-fibrogenic/pro-inflammatory *For correspondence: -

Glucagon-Like Peptide-1 Receptor Agonists Increase Pancreatic Mass by Induction of Protein Synthesis
Page 1 of 73 Diabetes Glucagon-like peptide-1 receptor agonists increase pancreatic mass by induction of protein synthesis Jacqueline A. Koehler1, Laurie L. Baggio1, Xiemin Cao1, Tahmid Abdulla1, Jonathan E. Campbell, Thomas Secher2, Jacob Jelsing2, Brett Larsen1, Daniel J. Drucker1 From the1 Department of Medicine, Tanenbaum-Lunenfeld Research Institute, Mt. Sinai Hospital and 2Gubra, Hørsholm, Denmark Running title: GLP-1 increases pancreatic protein synthesis Key Words: glucagon-like peptide 1, glucagon-like peptide-1 receptor, incretin, exocrine pancreas Word Count 4,000 Figures 4 Tables 1 Address correspondence to: Daniel J. Drucker M.D. Lunenfeld-Tanenbaum Research Institute Mt. Sinai Hospital 600 University Ave TCP5-1004 Toronto Ontario Canada M5G 1X5 416-361-2661 V 416-361-2669 F [email protected] 1 Diabetes Publish Ahead of Print, published online October 2, 2014 Diabetes Page 2 of 73 Abstract Glucagon-like peptide-1 (GLP-1) controls glucose homeostasis by regulating secretion of insulin and glucagon through a single GLP-1 receptor (GLP-1R). GLP-1R agonists also increase pancreatic weight in some preclinical studies through poorly understood mechanisms. Here we demonstrate that the increase in pancreatic weight following activation of GLP-1R signaling in mice reflects an increase in acinar cell mass, without changes in ductal compartments or β-cell mass. GLP-1R agonists did not increase pancreatic DNA content or the number of Ki67+ cells in the exocrine compartment, however pancreatic protein content was increased in mice treated with exendin-4 or liraglutide. The increased pancreatic mass and protein content was independent of cholecystokinin receptors, associated with a rapid increase in S6 kinase phosphorylation, and mediated through the GLP-1 receptor. -
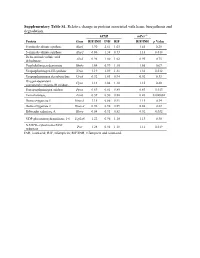
Supplementary Table S1. Relative Change in Proteins Associated with Heme Biosynthesis and Degradation
Supplementary Table S1. Relative change in proteins associated with heme biosynthesis and degradation. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value 5-aminolevulinate synthase Alas1 1.90 2.61 1.05 1.41 0.28 5-aminolevulinate synthase Alas2 0.86 1.38 0.73 1.18 0.018 Delta-aminolevulinic acid Alad 0.96 1.00 1.02 0.95 0.75 dehydratase Porphobilinogen deaminase Hmbs 1.04 0.99 1.10 1.05 0.67 Uroporphyrinogen-III synthase Uros 1.19 1.09 1.31 1.38 0.012 Uroporphyrinogen decarboxylase Urod 0.92 1.03 0.94 0.92 0.33 Oxygen-dependent Cpox 1.13 1.04 1.18 1.15 0.20 coproporphyrinogen-III oxidase, Protoporphyrinogen oxidase Ppox 0.69 0.81 0.85 0.83 0.013 Ferrochelatase, Fech 0.39 0.50 0.88 0.43 0.000002 Heme oxygenase 1 Hmox1 1.15 0.86 0.91 1.11 0.34 Heme oxygenase 2 Hmox2 0.96 0.98 0.89 0.88 0.22 Biliverdin reductase A Blvra 0.84 0.92 0.82 0.92 0.032 UDP-glucuronosyltransferase 1-6 Ugt1a6 1.22 0.96 1.10 1.13 0.30 NADPH--cytochrome P450 Por 1.28 0.92 1.18 1.12 0.019 reductase INH, isoniazid; RIF, rifampicin; RIF/INH, rifampicin and isoniazid. Supplementary Table S2. Relative change in protein nuclear receptors. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value Aryl hydrocarbon receptor Ahr 1.09 0.91 1.00 1.26 0.092 Hepatocyte nuclear factor Hnf1a 0.87 0.97 0.82 0.79 0.027 1-alpha Hepatocyte nuclear factor Hnf4a 0.95 1.05 0.97 1.08 0.20 4-alpha Oxysterols receptor LXR- Nr1h3 0.94 1.16 1.03 1.02 0.42 alpha Bile acid receptor Nr1h4 1.05 1.17 0.98 1.19 0.12 Retinoic acid receptor Rxra 0.88 1.03 0.83 0.95 0.12 RXR-alpha Peroxisome proliferator- -

The Expression Profile and Prognostic Significance of Eukaryotic Translation Elongation Factors in Different Cancers
RESEARCH ARTICLE The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers Md. Khurshidul Hassan, Dinesh Kumar, Monali Naik, Manjusha Dixit* School of Biological Sciences, National Institute of Science Education and Research, HBNI, Bhimpur- Padanpur, Jatni, Khurda, Odisha, India * [email protected] a1111111111 Abstract a1111111111 Eukaryotic translation factors, especially initiation factors have garnered much attention a1111111111 a1111111111 with regards to their role in the onset and progression of different cancers. However, the a1111111111 expression levels and prognostic significance of translation elongation factors remain poorly explored in different cancers. In this study, we have investigated the mRNA transcript levels of seven translation elongation factors in different cancer types using Oncomine and TCGA databases. Furthermore, we have identified the prognostic significance of these factors OPEN ACCESS using Kaplan-Meier Plotter and SurvExpress databases. We observed altered expression levels of all the elongation factors in different cancers. Higher expression of EEF1A2, Citation: Hassan MK, Kumar D, Naik M, Dixit M (2018) The expression profile and prognostic EEF1B2, EEF1G, EEF1D, EEF1E1 and EEF2 was observed in most of the cancer types, significance of eukaryotic translation elongation whereas reverse trend was observed for EEF1A1. Overexpression of many factors pre- factors in different cancers. PLoS ONE 13(1): dicted poor prognosis in breast (EEF1D, EEF1E1, EEF2) and lung cancer (EEF1A2, e0191377. https://doi.org/10.1371/journal. EEF1B2, EEF1G, EEF1E1). However, we didn't see any common correlation of expression pone.0191377 levels of elongation factors with survival outcomes across cancer types. Cancer subtype Editor: Aamir Ahmad, University of South Alabama stratification showed association of survival outcomes and expression levels of elongation Mitchell Cancer Institute, UNITED STATES factors in specific sub-types of breast, lung and gastric cancer.