Oenothera, a Unique Model to Study the Role of Plastids in Speciation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Gigas Effect: a Reliable Predictor of Ploidy? Case Studies in Oxalis
The gigas effect: A reliable predictor of ploidy? Case studies in Oxalis by Frederik Willem Becker Thesis presented in fulfilment of the requirements for the degree of Master of Science in the Faculty of Science at Stellenbosch University Supervisors: Prof. Léanne L.Dreyer, Dr. Kenneth C. Oberlander, Dr. Pavel Trávníček March 2021 Stellenbosch University https://scholar.sun.ac.za Declaration By submitting this thesis electronically, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof (save to the extent explicitly otherwise stated), that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. March 2021 …………………………. ………………… F.W. Becker Date Copyright © 2021 Stellenbosch University All rights reserved i Stellenbosch University https://scholar.sun.ac.za Abstract Whole Genome Duplication (WGD), or polyploidy is an important evolutionary process, but literature is divided over its long-term evolutionary potential to generate diversity and lead to lineage divergence. WGD often causes major phenotypic changes in polyploids, of which the most prominent is the Gigas effect. The Gigas effect refers to the enlargement of plant cells due to their increased amount of DNA, causing plant organs to enlarge as well. This enlargement has been associated with fitness advantages in polyploids, enabling them to successfully establish and persist, eventually causing speciation. Using Oxalis as a study system, I examine whether Oxalis polyploids exhibit the Gigas effect using 24 species across the genus from the Oxalis living research collection at the Stellenbosch University Botanical Gardens, Stellenbosch. -

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Differences in Copper Absorption and Accumulation Between Copper-Exclusion and Copper-Enrichment Plants: a Comparison of Structure and Physiological Responses
RESEARCH ARTICLE Differences in Copper Absorption and Accumulation between Copper-Exclusion and Copper-Enrichment Plants: A Comparison of Structure and Physiological Responses Lei Fu, Chen Chen, Bin Wang, Xishi Zhou, Shuhuan Li, Pan Guo, Zhenguo Shen, Guiping Wang, Yahua Chen* College of Life Sciences, Jiangsu Collaborative Innovation Center for Solid Organic Waste Resource, National Joint Local Engineering Research Center for Rural Land Resources Use and Consolidation, Nanjing Agricultural University, Nanjing, Jiangsu, China * [email protected] OPEN ACCESS Abstract Citation: Fu L, Chen C, Wang B, Zhou X, Li S, Guo Differences in copper (Cu) absorption and transport, physiological responses and structural P, et al. (2015) Differences in Copper Absorption and characteristics between two types of Cu-resistant plants, Oenothera glazioviana (Cu-exclu- Accumulation between Copper-Exclusion and sion type) and Elsholtzia haichowensis (Cu-enrichment type), were investigated in the pres- Copper-Enrichment Plants: A Comparison of Structure and Physiological Responses. PLoS ONE ent study. The results indicated the following: (1) After 50 μM Cu treatment, the Cu ratio in 10(7): e0133424. doi:10.1371/journal.pone.0133424 the xylem vessels of E. haichowensis increased by 60%. A Cu adsorption experiment Editor: Wei Wang, Henan Agricultural Univerisity, indicated that O. glazioviana exhibited greater resistance to Cu, and Cu absorption and the CHINA shoot/root ratio of Cu were significantly lower in O. glazioviana than in E. haichowensis. (2) Received: April 28, 2015 An analysis of the endogenous abscisic acid (ABA) variance and exogenous ABA treatment demonstrated that the ABA levels of both plants did not differ; exogenous ABA treatment Accepted: June 26, 2015 clearly reduced Cu accumulation in both plants. -
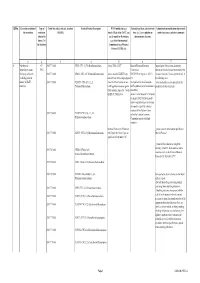
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Terrestrial and Marine Biological Resource Information
APPENDIX C Terrestrial and Marine Biological Resource Information Appendix C1 Resource Agency Coordination Appendix C2 Marine Biological Resources Report APPENDIX C1 RESOURCE AGENCY COORDINATION 1 The ICF terrestrial biological team coordinated with relevant resource agencies to discuss 2 sensitive biological resources expected within the terrestrial biological study area (BSA). 3 A summary of agency communications and site visits is provided below. 4 California Department of Fish and Wildlife: On July 30, 2020, ICF held a conference 5 call with Greg O’Connell (Environmental Scientist) and Corianna Flannery (Environmental 6 Scientist) to discuss Project design and potential biological concerns regarding the 7 Eureka Subsea Fiber Optic Cables Project (Project). Mr. O’Connell discussed the 8 importance of considering the western bumble bee. Ms. Flannery discussed the 9 importance of the hard ocean floor substrate and asked how the cable would be secured 10 to the ocean floor to reduce or eliminate scour. The western bumble bee has been 11 evaluated in the Biological Resources section of the main document, and direct and 12 indirect impacts are avoided. The Project Description describes in detail how the cable 13 would be installed on the ocean floor, the importance of the hard bottom substrate, and 14 the need for avoidance. 15 Consultation Outcomes: 16 • The Project was designed to avoid hard bottom substrate, and RTI Infrastructure 17 (RTI) conducted surveys of the ocean floor to ensure that proper routing of the 18 cable would occur. 19 • Ms. Flannery will be copied on all communications with the National Marine 20 Fisheries Service 21 California Department of Fish and Wildlife: On August 7, 2020, ICF held a conference 22 call with Greg O’Connell to discuss a site assessment and survey approach for the 23 western bumble bee. -

National List of Vascular Plant Species That Occur in Wetlands 1996
National List of Vascular Plant Species that Occur in Wetlands: 1996 National Summary Indicator by Region and Subregion Scientific Name/ North North Central South Inter- National Subregion Northeast Southeast Central Plains Plains Plains Southwest mountain Northwest California Alaska Caribbean Hawaii Indicator Range Abies amabilis (Dougl. ex Loud.) Dougl. ex Forbes FACU FACU UPL UPL,FACU Abies balsamea (L.) P. Mill. FAC FACW FAC,FACW Abies concolor (Gord. & Glend.) Lindl. ex Hildebr. NI NI NI NI NI UPL UPL Abies fraseri (Pursh) Poir. FACU FACU FACU Abies grandis (Dougl. ex D. Don) Lindl. FACU-* NI FACU-* Abies lasiocarpa (Hook.) Nutt. NI NI FACU+ FACU- FACU FAC UPL UPL,FAC Abies magnifica A. Murr. NI UPL NI FACU UPL,FACU Abildgaardia ovata (Burm. f.) Kral FACW+ FAC+ FAC+,FACW+ Abutilon theophrasti Medik. UPL FACU- FACU- UPL UPL UPL UPL UPL NI NI UPL,FACU- Acacia choriophylla Benth. FAC* FAC* Acacia farnesiana (L.) Willd. FACU NI NI* NI NI FACU Acacia greggii Gray UPL UPL FACU FACU UPL,FACU Acacia macracantha Humb. & Bonpl. ex Willd. NI FAC FAC Acacia minuta ssp. minuta (M.E. Jones) Beauchamp FACU FACU Acaena exigua Gray OBL OBL Acalypha bisetosa Bertol. ex Spreng. FACW FACW Acalypha virginica L. FACU- FACU- FAC- FACU- FACU- FACU* FACU-,FAC- Acalypha virginica var. rhomboidea (Raf.) Cooperrider FACU- FAC- FACU FACU- FACU- FACU* FACU-,FAC- Acanthocereus tetragonus (L.) Humm. FAC* NI NI FAC* Acanthomintha ilicifolia (Gray) Gray FAC* FAC* Acanthus ebracteatus Vahl OBL OBL Acer circinatum Pursh FAC- FAC NI FAC-,FAC Acer glabrum Torr. FAC FAC FAC FACU FACU* FAC FACU FACU*,FAC Acer grandidentatum Nutt. -

New Records of Vascular Plant Distribution in the Polish Part of the Lithuanian Lakeland, North-Eastern Poland
10.2478/botlit-2019-0011 BOTANICA ISSN 2538-8657 2019, 25(1): 97–101 Short note NEW RECORDS OF VASCULAR PLANT DISTRIBUTION IN THE POLISH PART OF THE LITHUANIAN LAKELAND, NORTH-EASTERN POLAND Artur PLISZKO 1* & Monika Wo ź n i a k -Ch o d a C k a 2 1 Jagiellonian University, Institute of Botany, Department of Taxonomy, Phytogeography and Palaeobotany, Gronostajowa Str. 3, Kraków PL-30-387, Poland 2 Polish Academy of Sciences, W. Szafer Institute of Botany, Department of Vascular Plants, Lubicz Str. 46, Kraków PL-31-512, Poland *Corresponding author. E-mail: [email protected] Abstract Pliszko A., Woźniak-Chodacka M., 2019: New records of vascular plant distribution in the Polish part of the Lithuanian Lakeland, north-eastern Poland. – Botanica, 25(1): 97–101. The paper presents new localities of 15 vascular plants recorded in the Polish part of the Lithuanian Lake- land, north-eastern Poland in 2017–2018, using the ATPOL cartogram method. Crepis capillaris, Diplotaxis tenuifolia, Eragrostis albensis, Matthiola longipetala, Oenothera fruticosa, Oenothera glazioviana and Rubus armeniacus are listed as new species for the regional flora. Keywords: alien species, biological recording, floristics, geographical distribution, vascular plants. In many regions of Poland, distribution of vascular 20th century (so K o ł o w s K i , 1965, 1973, 1988a, 1988b; plants is insufficiently recognized due to a lack of in- MAZUR et al., 1978). The flora of the Polish part of tensive floristic studies (Za j ą c & Za j ą c , 2001, 2015). the Lithuanian Lakeland still attracts the interest The Lithuanian Lakeland is a physical-geographical among researchers (ja b ł o ń s K a , 2005; Pa w L i K o w s K i , macroregion between Poland, Russia (Kaliningrad 2008a, 2008b; Pa w L i K o w s K i et al., 2011, 2013; Oblast), Lithuania and Belarus. -

Hybridization Between a Rare and Introduced Oenothera Along the North Pacific Coast
Western North American Naturalist Volume 68 Number 2 Article 4 6-10-2008 Hybridization between a rare and introduced Oenothera along the north Pacific Coast Matthew L. Carlson University of Alaska, Anchorage Robert J. Meinke Oregon State University, Corvallis Follow this and additional works at: https://scholarsarchive.byu.edu/wnan Recommended Citation Carlson, Matthew L. and Meinke, Robert J. (2008) "Hybridization between a rare and introduced Oenothera along the north Pacific Coast," Western North American Naturalist: Vol. 68 : No. 2 , Article 4. Available at: https://scholarsarchive.byu.edu/wnan/vol68/iss2/4 This Article is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Western North American Naturalist by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Western North American Naturalist 68(2), © 2008, pp. 161–172 HYBRIDIZATION BETWEEN A RARE AND INTRODUCED OENOTHERA ALONG THE NORTH PACIFIC COAST Matthew L. Carlson1 and Robert J. Meinke2 ABSTRACT.—Interspecific hybridization has increasingly become regarded as a serious threat to the genetic integrity and persistence of rare plants. Oenothera glazioviana (Onagraceae) is a horticultural species that has escaped cultivation and now threatens the narrow Pacific coastal endemic O. wolfii with hybridization. Reports of morphologically intermediate and ecologically aggressive forms prompted this investigation into the extent of hybridization over the range of O. wolfii. In particular, this study identifies populations of pure and hybrid origins. We used multivariate methods to characterize the morphological variation of Oregon and northern California coastal Oenothera populations. -

17 December 2014 FIELD CHECKLIST of the VASCULAR
17 December 2014 TAXACEAE (Yew Family) __ Taxus canadensis Canada Yew FIELD CHECKLIST OF THE VASCULAR PLANTS OF JOKERS HILL King Township, Regional Municipality of York PINACEAE (Pine Family) C.S Blaney and P.M. Kotanen* __ Larix laricina Tamarack *Correspondence author: __ Picea glauca White Spruce Department of Ecology & Evolutionary Biology __ Pinus strobus White Pine University of Toronto at Mississauga __ Pinus sylvestris Scots Pine 3359 Mississauga Road North __ Tsuga canadensis Eastern Hemlock Mississauga, ON Canada CUPRESSACEAE (Cypress Family) e-mail: [email protected] __ Juniperus communis Common Juniper __ Juniperus virginiana Eastern Red Cedar The following list is based on observations and collections by the authors between 1997 and 1999, __ Thuja occidentalis Eastern White Cedar with later additions by numerous observers. It includes all species known to be growing outside of cultivation within the Jokers Hill property. Also listed are native species found in land adjacent to the TYPHACEAE (Cat-tail Family) Jokers Hill property, but not yet found on the site (scientific name preceded by "*" - 13 species). A __ Typha angustifolia Narrow-leaved Cat-tail total of 631 taxa (species and hybrids) are listed; 450 taxa are considered native to Jokers Hill (those __ Typha latifolia Common Cat-tail listed in bold typeface) and 181 are considered non-native (listed in regular typeface). Determining native versus non-native status required a few rather arbitrary judgements. SPARGANIACEAE (Bur-reed Family) __ Sparganium chlorocarpum Green Bur-reed Several people assisted in the preparation of this list. P. Ball of the University of Toronto at Mississauga and A. -

Buchbesprechungen 247-296 ©Verein Zur Erforschung Der Flora Österreichs; Download Unter
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Neilreichia - Zeitschrift für Pflanzensystematik und Floristik Österreichs Jahr/Year: 2006 Band/Volume: 4 Autor(en)/Author(s): Mrkvicka Alexander Ch., Fischer Manfred Adalbert, Schneeweiß Gerald M., Raabe Uwe Artikel/Article: Buchbesprechungen 247-296 ©Verein zur Erforschung der Flora Österreichs; download unter www.biologiezentrum.at Neilreichia 4: 247–297 (2006) Buchbesprechungen Arndt KÄSTNER, Eckehart J. JÄGER & Rudolf SCHUBERT, 2001: Handbuch der Se- getalpflanzen Mitteleuropas. Unter Mitarbeit von Uwe BRAUN, Günter FEYERABEND, Gerhard KARRER, Doris SEIDEL, Franz TIETZE, Klaus WERNER. – Wien & New York: Springer. – X + 609 pp.; 32 × 25 cm; fest gebunden. – ISBN 3-211-83562-8. – Preis: 177, – €. Dieses imposante Kompendium – wohl das umfangreichste Werk zu diesem Thema – behandelt praktisch alle Aspekte der reinen und angewandten Botanik rund um die Ackerbeikräuter. Es entstand in der Hauptsache aufgrund jahrzehntelanger Forschungs- arbeiten am Institut für Geobotanik der Universität Halle über Ökologie und Verbrei- tung der Segetalpflanzen. Im Zentrum des Werkes stehen 182 Arten, die ausführlich behandelt werden, wobei deren eindrucksvolle und umfassende „Porträt-Zeichnungen“ und genaue Verbreitungskarten am wichtigsten sind. Der „Allgemeine“ Teil („I.“) beginnt mit der Erläuterung einiger (vor allem morpholo- gischer, ökologischer, chorologischer und zoologischer) Fachausdrücke, darauf -

Pelargoniums an Herb Society of America Guide
Pelargoniums An Herb Society of America Guide The Herb Society of America 9019 Kirtland Chardon Rd. Kirtland, Ohio 44094 © 2006 The Herb Society of America Pelargoniums: An Herb Society of America Guide Table of Contents Introduction …………………………………………………………….…. 3 Contributors & Acknowledgements ……………………………………… 3 Description & Taxonomy ..………………………………………………... 8 Chemistry …………………………………………………………………. 10 Nutrition …………………………………………………………………... 10 History & Folklore ………………………………………………………… 10 Literature & Art …………………………………………………………… 12 Cultivation ………………………………………………………………… 13 Pests & Diseases …………………………………………………………... 19 Pruning & Harvesting ……………………………………………………… 20 Preserving & Storing ………………………………………………………. 21 Uses ………………………………………………………………………... 21 - Culinary Uses ………………………………………………… 21 - Recipes ………………………………………………… 23 - Craft Uses ……………………………………………………. 40 - Cosmetic Uses ……………………………………………….. 41 - Recipes ……………………………………………….. 42 - Medicinal & Ethnobotanical Uses & Aromatherapy ………... 43 - Garden Uses ………………………………………………….. 47 - Other Uses …………………………………………………... 48 Species Highlights …..……………………………………………………… 49 Cultivar Examples …………………………………………………………. 57 Literature Citations & References ………………………………………... 62 HSA Library Pelargonium Resources …...………………………………… 68 © The Herb Society of America - 9019 Kirtland Chardon Rd., Kirtland, OH, 44094 - (440) 256-0514 - http://www.herbsociety.org 2 Pelargoniums: An Herb Society of America Guide Introduction Mission: The Herb Society of America is dedicated to promoting the knowledge, use and delight of