Molecular Cloning and Structural Modelling of Gamma-Phospholipase A2 Inhibitors from Bothrops Atrox and Micrurus Lemniscatus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Endogenous Phospholipase A2 Inhibitors in Snakes: a Brief Overview
Campos et al. Journal of Venomous Animals and Toxins including Tropical Diseases (2016) 22:37 DOI 10.1186/s40409-016-0092-5 REVIEW Open Access Endogenous phospholipase A2 inhibitors in snakes: a brief overview Patrícia Cota Campos†, Lutiana Amaral de Melo†, Gabriel Latorre Fortes Dias and Consuelo Latorre Fortes-Dias* Abstract The blood plasma of numerous snake species naturally comprises endogenous phospholipase A2 inhibitors, which primarily neutralize toxic phospholipases A2 that may eventually reach their circulation. This inhibitor type is generally known as snake blood phospholipase A2 inhibitors (sbPLIs). Most, if not all sbPLIs are oligomeric glycosylated proteins, although the carbohydrate moiety may not be essential for PLA2 inhibition in every case. The presently known sbPLIs belong to one of three structural classes – namely sbαPLI, sbβPLI or sbγPLI – depending on the presence of characteristic C-type lectin-like domains, leucine-rich repeats or three-finger motifs, respectively. Currently, the most numerous inhibitors described in the literature are sbαPLIs and sbγPLIs, whereas sbβPLIs are rare. When the target PLA2 is a Lys49 homolog or an Asp49 myotoxin, the sbPLI is denominated a myotoxin inhibitor protein (MIP). In this brief overview, the most relevant data on sbPLIs will be presented. Representative examples of sbαPLIs and sbγPLIs from two Old World – Gloydius brevicaudus and Malayopython reticulatus – and two New World – Bothrops alternatus and Crotalus durissus terrificus – snake species will be emphasized. Keywords: PLA2 inhibitor, Phospholipase A2, Snake blood, Natural resistance, Snakes Background domain of Ca2+-dependent lectins, and preferentially in- A number of venomous and nonvenomous snake species hibit acidic PLA2s. Beta-type inhibitors (sbβPLIs) exhibit are naturally resistant to the deleterious actions of snake tandem leucine-rich repeats (LRRs), and specifically inhibit venom components, in many cases due to the presence basic PLA2s. -

Azure-Winged Magpie Onaga (Jpn) Cyanopica Cyana
Bird Research News Vol.6 No.6 2009.6.24. Azure-winged Magpie Onaga (Jpn) Cyanopica cyana Morphology and classification Flock: Azure-winged Magpies live in a flock in the breeding and non- Classification: Passeriformes Corvidae breeding seasons, holding their flock territory throughout the year (Hosono 1989). In breeding period they roost in a flock except for Total length: 366.8mm (319-390) Wing length: 130.7mm (122-141) females incubating eggs and nestlings. In Nagano Pref., for instance, Tail length: 214.8mm (192-240) Culmen length: 25.7mm (24-30) the mean flock and home range sizes were 23 birds (9-45) and 21.8 ha Tarsus length: 33.3mm (32-35) Weight: 83.4g (69-96) (11-48), respectively in Kawanakajima (Hosono 1968), 28.7 birds and 135.1ha (103-243) in Ina, and 16.7 birds and 287.6 ha (130-376) in Measurements by Kuzu (1942). Nobeyama (Imanishi 2003). In Saitama Pref., on the other hand, they Appearance: were 24 birds (17-31, n = 16) and 13.4ha (6.2-24.8, n = 11) respec- Azure-winged Magpies are similar in tively in Tokorozawa, where Azure-winged Magpies are assumed to plumage coloration in males and fe- occur in the highest density. They also roost in a flock, but more than one flock occasionally roosted together in the same site. They use as a males. Males are slightly larger than roost site a dense thicket of bamboo, a coniferous wood and a broad- females in body size. They are gray on leaved deciduous wood. A coniferous wood and a thicket of bamboo the upperpart and white or light gray were used with higher frequency in winter, but a broad-leaved decidu- on the underpart (Photo 1). -

A Rapid Survey of Online Trade in Live Birds and Reptiles in The
S H O R T R E P O R T 0ൾඍඁඈൽඌ A rapid online survey was undertaken EHWZHHQDQG)HEUXDU\ GD\V DSSUR[LPDWHO\KRXUVVXUYH\GD\ RQ pre-selected Facebook groups specializing in the trade of live pets. Ten groups each for reptiles and birds were selected based on trading activities in the previous six months. The survey was carried out during ZHHN GD\V 0RQGD\ WR )ULGD\ E\ JRLQJ through each advertisement posted in A rapid survey of online trade in the groups. Information, including that live birds and reptiles in the Philippines relating to species, quantity, and asking HYDROSAURUS PUSTULATUS WWF / URS WOY WOY WWF / URS PUSTULATUS HYDROSAURUS SULFH ZDV QRWHG 6SHFLHV ZHUH LGHQWL¿HG Report by Cristine P. Canlas, Emerson Y. Sy, to the lowest taxonomic level whenever and Serene Chng possible. Taxonomy follows Gill and 'RQVNHU IRU ELUGV DQG 8HW] et al. IRUUHSWLOHV7KHDXWKRUVFDOFXODWHG ,ඇඍඋඈൽඎർඍංඈඇ WKH WRWDO SRWHQWLDO YDOXH R൵HUHG IRU ELUGV and reptiles based on prices indicated he Philippines is the second largest archipelago in the world by traders. Advertisements that did not comprising 7641 islands and is both a mega-biodiverse specify prices were assigned the lowest country for harbouring wildlife species found nowhere known price for each taxon. Valuations in else in the world, and one of eight biodiversity hotspots this report were based on a conversion rate having a disproportionate number of species threatened with RI86' 3+3 $QRQ ,WLV ,//8675$7,213+,/,33,1(6$,/),1/,=$5' TH[WLQFWLRQIXUWKHULWKDVVRPHRIWKHKLJKHVWUDWHVRIHQGHPLFLW\LQWKH not always possible during online surveys world (Myers et al 7KHLOOHJDOZLOGOLIHWUDGHLVRQHRIWKHPDLQ WRYHULI\WKDWDOOR൵HUVDUHJHQXLQH UHDVRQVEHKLQGVLJQL¿FDQWGHFOLQHVRIVRPHZLOGOLIHSRSXODWLRQVLQ$VLD LQFOXGLQJWKH3KLOLSSLQHV $QRQ6RGKLet al1LMPDQDQG 5ൾඌඎඅඍඌ 6KHSKHUG'LHVPRVet al5DRet al 7KHWildlife Act of 2001 (Republic Act No. -

Prey-Handling Behavior of Hatchling Elaphe Helena (Colubridae)
Herpetologica, 59(4), 2003, 469–474 Ó 2003 by The Herpetologists’ League, Inc. PREY-HANDLING BEHAVIOR OF HATCHLING ELAPHE HELENA (COLUBRIDAE) 1,2 RITA S. MEHTA Department of Biology, University of Texas, Tyler, TX 75719, USA ABSTRACT: The effects of prey size on prey-handling behavior for 60 ingestively naive hatchling Elaphe helena were studied in the laboratory. Hatchlings were randomly assigned to one of three diet categories in which prey (Mus musculus) varied by relative mass differences of 20–35%, 40–46%, or 50–59% of an individual snake’s own body mass. The effects of prey size on capture position, direction of ingestion, condition of prey at ingestion (dead/alive), feeding duration, and prey-handling tactic were observed and recorded for each feeding episode. Results indicated that prey size significantly affected the prey-handling behavior of hatchling E. helena. In the largest relative mass category, hatchlings captured prey by the anterior end more often than in the smaller two relative mass categories. Prey from the smallest relative mass category were simply seized whereas, in the medium and large categories, pinion and constriction behaviors were observed. Time to subdue and ingest the prey item increased with prey size categories. Key words: Colubridae; Effects of prey size; Elaphe helena; Prey-handling behavior THE SIZE, type, and activity level of various (i.e., press prey against the substrate with the prey are thought to influence the feeding anterior portion of the body) and constrict behaviors for many advanced snakes (de small active mice than nestling rats. Further- Queiroz, 1984; Moon, 2000). -

Risk Analysis of the Russian Rat Snake ( ) in the Netherlands Elaphe Schrenckii
RISK ANALYSIS OF THE RUSSIAN RAT SNAKE (ELAPHE SCHRENCKII ) IN THE NETHERLANDS Commissioned by: Invasive Alien Species Team Netherlands Food and Consumer Product Safety Authority Ministry of Economic Affairs, Agriculture and Innovation RISK ANALYSIS OF THE RUSSIAN RAT SNAKE (ELAPHE SCHRENCKII) IN THE NETHERLANDS S. van de Koppel MSc drs. N. van Kessel ir. B.H.J.M. Crombaghs W. Getreuer dr. H.J.R. Lenders Commissioned by: Invasive Alien Species Team Netherlands Food and Consumer Product Safety Authority Ministry of Economic Affairs, Agriculture and Innovation 2012-04-25 2012 Natuurbalans Limes-Divergens BV Text and composition: S. van de Koppel MSc1, drs. N. van Kessel 1, ir. B.H.J.M. Crombaghs1, W. Getreuer2, dr. H.J.R. Lenders3 1 Natuurbalans-Limes Divergens BV, Nijmegen, the Netherlands 2 ReptielenZoo SERPO, Delft, the Netherlands 3 Radboud University, Nijmegen, the Netherlands Signed for publication: Managing Director, Natuurbalans-Limes Divergens BV, Nijmegen, the Netherlands ir. B.H.J.M. Crombaghs Project code: 11-131 Commissioned by: ir. J.W. Lammers Invasive Alien Species Team, Netherlands Food and Consumer Product Safety Authority, Ministry of Economic Affairs, Agriculture and Innovation (Team Invasieve Exoten, Nederlandse Voedsel en Waren Autoriteit, Ministerie van Economische Zaken, Landbouw en Innovatie) Cover photos: W. Getreuer, S. van de Koppel Citation: Van de Koppel, S., N. van Kessel, B.H.J.M. Crombaghs, W. Getreuer & H.J.R. Lenders, 2012. Risk Analysis of the Russian Rat Snake (Elaphe schrenckii) in the Netherlands. Natuurbalans - Limes Divergens BV, Nijmegen / ReptielenZoo SERPO, Delft / Radboud University, Nijmegen. No part of this report may be reproduced and/or published by means of scanning, internet, photocopy, microfilm or any other means, without the prior written consent of the client indicated above and Natuurbalans-Limes Divergens BV nor may it, without such approval, be used for any work other than for which it was manufactured. -

The Wide Distribution and Change of Target Specificity of R2 Non-LTR Retrotransposons in Animals
RESEARCH ARTICLE The Wide Distribution and Change of Target Specificity of R2 Non-LTR Retrotransposons in Animals Kenji K. Kojima1,2,3*, Yosuke Seto2¤, Haruhiko Fujiwara2 1 Genetic Information Research Institute, Mountain View, CA, 94043, United States of America, 2 Graduate School of Frontier Sciences, University of Tokyo, Kashiwa, Chiba, 277±8562, Japan, 3 Department of Life Sciences, National Cheng Kung University, Tainan, 701, Taiwan a11111 ¤ Current address: Tokyo Metropolitan University, Hachioji, Tokyo, 192±0397, Japan * [email protected] Abstract Transposons, or transposable elements, are the major components of genomes in most OPEN ACCESS eukaryotes. Some groups of transposons have developed target specificity that limits the Citation: Kojima KK, Seto Y, Fujiwara H (2016) The integration sites to a specific nonessential sequence or a genomic region to avoid gene dis- Wide Distribution and Change of Target Specificity ruption caused by insertion into an essential gene. R2 is one of the most intensively investi- of R2 Non-LTR Retrotransposons in Animals. PLoS ONE 11(9): e0163496. doi:10.1371/journal. gated groups of sequence-specific non-LTR retrotransposons and is inserted at a specific pone.0163496 site inside of 28S ribosomal RNA (rRNA) genes. R2 is known to be distributed among at Editor: Ruslan Kalendar, University of Helsinki, least six animal phyla even though its occurrence is reported to be patchy. Here, in order to FINLAND obtain a more detailed picture of the distribution of R2, we surveyed R2 using both in silico Received: May 17, 2016 screening and degenerate PCR, particularly focusing on actinopterygian fish. We found two families of the R2C lineage from vertebrates, although it has previously only been Accepted: September 9, 2016 found in platyhelminthes. -

Captive Wildlife Regulations, 2021, W-13.12 Reg 5
1 CAPTIVE WILDLIFE, 2021 W-13.12 REG 5 The Captive Wildlife Regulations, 2021 being Chapter W-13.12 Reg 5 (effective June 1, 2021). NOTE: This consolidation is not official. Amendments have been incorporated for convenience of reference and the original statutes and regulations should be consulted for all purposes of interpretation and application of the law. In order to preserve the integrity of the original statutes and regulations, errors that may have appeared are reproduced in this consolidation. 2 W-13.12 REG 5 CAPTIVE WILDLIFE, 2021 Table of Contents PART 1 PART 5 Preliminary Matters Zoo Licences and Travelling Zoo Licences 1 Title 38 Definition for Part 2 Definitions and interpretation 39 CAZA standards 3 Application 40 Requirements – zoo licence or travelling zoo licence PART 2 41 Breeding and release Designations, Prohibitions and Licences PART 6 4 Captive wildlife – designations Wildlife Rehabilitation Licences 5 Prohibition – holding unlisted species in captivity 42 Definitions for Part 6 Prohibition – holding restricted species in captivity 43 Standards for wildlife rehabilitation 7 Captive wildlife licences 44 No property acquired in wildlife held for 8 Licence not required rehabilitation 9 Application for captive wildlife licence 45 Requirements – wildlife rehabilitation licence 10 Renewal 46 Restrictions – wildlife not to be rehabilitated 11 Issuance or renewal of licence on terms and conditions 47 Wildlife rehabilitation practices 12 Licence or renewal term PART 7 Scientific Research Licences 13 Amendment, suspension, -

Uitheemse Slangen in Nederland
Alterra Wageningen UR Alterra Wageningen UR is hét kennisinstituut voor de groene leefomgeving en Postbus 47 bundelt een grote hoeveelheid expertise op het gebied van de groene ruimte en het Uitheemse slangen in Nederland 6700 AA Wageningen duurzaam maatschappelijk gebruik ervan: kennis van water, natuur, bos, milieu, T 317 48 07 00 bodem, landschap, klimaat, landgebruik, recreatie etc. www.wageningenUR.nl/alterra De missie van Wageningen UR (University & Research centre) is ‘To explore the potential of nature to improve the quality of life’. Binnen Wageningen UR bundelen Alterra-rapport 2496 9 gespecialiseerde onderzoeksinstituten van stichting DLO en Wageningen ISSN 1566-7197 Een analyse van de kans op introductie, vestiging, uitbreiding en schade University hun krachten om bij te dragen aan de oplossing van belangrijke vragen in het domein van gezonde voeding en leefomgeving. Met ongeveer 30 vestigingen, 6.000 medewerkers en 9.000 studenten behoort Wageningen UR wereldwijd tot de R.J.F. Bugter, S. van de Koppel, R.C.M. Creemers, A.J. Griffioen en F.G.W.A. Ottburg aansprekende kennisinstellingen binnen haar domein. De integrale benadering van de vraagstukken en de samenwerking tussen verschillende disciplines vormen het hart van de unieke Wageningen aanpak. Uitheemse slangen in Nederland Een analyse van de kans op introductie, vestiging, uitbreiding en schade R.J.F. Bugter1, S. van de Koppel2, R.C.M. Creemers3, A.J. Griffioen1 en F.G.W.A. Ottburg1 1 Alterra 2 Natuurbalans-Limes Divergens 3 RAVON Reptielen Amfibieën Vissen Onderzoek Nederland Dit onderzoek is uitgevoerd in opdracht van de Nederlandse Voedsel en Waren Autoriteit. Alterra Wageningen UR Wageningen, januari 2014 Alterra-rapport 2496 ISSN 1566-7197 RAVON-Rapport 2013.112 Natuurbalans-Limes Divergens-Rapport 12-181 Bugter, R.J.F., S. -
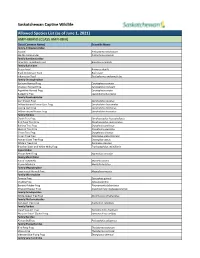
Captive Wildlife Allowed List
Saskatchewan Captive Wildlife Allowed Species List (as of June 1, 2021) AMPHIBIANS (CLASS AMPHIBIA) Class (Common Name) Scientific Name Family Ambystomatidae Axolotl Ambystoma mexicanum Marble Salamander Ambystoma opacum Family Bombinatoridae Oriental Fire-Bellied Toad Bombina orientalis Family Bufonidae Green Toad Anaxyrus debilis Black Indonesian Toad Bufo asper Indonesian Toad Duttaphrynus melanostictus Family Ceratophryidae Surinam Horned Frog Ceratophrys cornuta Chacoan Horned Frog Ceratophrys cranwelli Argentine Horned Frog Ceratophrys ornata Budgett’s Frog Lepidobatrachus laevis Family Dendrobatidae Dart Poison Frog Dendrobates auratus Yellow-banded Poison Dart Frog Dendrobates leucomelas Dyeing Dart Frog Dendrobates tinctorius Yellow-striped Poison Frog Dendrobates truncatus Family Hylidae Clown Tree Frog Dendropsophus leucophyllatus Bird Poop Tree Frog Dendropsophus marmoratus Barking Tree Frog Dryophytes gratiosus Squirrel Tree Frog Dryophytes squirellus Green Tree Frog Dryophytes cinereus Cuban Tree Frog Osteopilus septentrionalis Haitian Giant Tree Frog Osteopilus vastus White’s Tree Frog Ranoidea caerulea Brazilian Black and White Milky Frog Trachycephalus resinifictrix Hyperoliidae African Reed Frog Hyperolius concolor Family Mantellidae Baron’s Mantella Mantella baroni Brown Mantella Mantella betsileo Family Megophryidae Long-nosed Horned Frog Megophrys nasuta Family Microhylidae Tomato Frog Dyscophus guineti Chubby Frog Kaloula pulchra Banded Rubber Frog Phrynomantis bifasciatus Emerald Hopper Frog Scaphiophryne madagascariensis -

Zootaxa, at the Lower Size Limit in Snakes
Zootaxa 1841: 1–30 (2008) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2008 · Magnolia Press ISSN 1175-5334 (online edition) At the lower size limit in snakes: two new species of threadsnakes (Squamata: Leptotyphlopidae: Leptotyphlops) from the Lesser Antilles S. BLAIR HEDGES Department of Biology, Pennsylvania State University, 208 Mueller Laboratory, University Park, PA 16802-5301 USA. E-mail:[email protected] Abstract Islands are viewed as natural evolutionary laboratories for terrestrial organisms because they have boundaries that limit dispersal and often reveal evolutionary patterns and mechanisms. One such pattern is that the smallest and largest species of different types of tetrapod animals are frequently found on islands. Here I describe two new diminutive species of snakes of the genus Leptotyphlops from the Lesser Antilles: one from Saint Lucia and the other from Barbados. The one from Barbados is the smallest species of snake and has a total adult length of approximately 100 mm. Limited evidence indicates a clutch size of one and a greatly elongated egg shape (length /width). Comparison of egg shapes in snakes indi- cates that the shape is a packaging phenomenon, related primarily to the shape of the available body cavity and clutch size. For a clutch size of one, expected egg shape is eight whereas expected egg shape drops to two at a clutch size of ten. The body shape of snakes, defined as snout-to-vent length divided by width, also varies and influences the shape of snake eggs. The smallest snakes are typically stout-bodied with shapes of 30–35 whereas the longest snakes usually are more elongate, with shapes of 45–50. -

Globin Gene Families in Sauropsid Vertebrates Federico G
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications in the Biological Sciences Papers in the Biological Sciences 1-2018 Gene Turnover and Diversification of the α- and β- Globin Gene Families in Sauropsid Vertebrates Federico G. Hoffmann University of Nebraska-Lincoln, [email protected] Michael W. Vandewege Texas Tech University Jay F. Storz University of Nebraska - Lincoln, [email protected] Juan C. Opazo Universidad Austral de Chile, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/bioscifacpub Part of the Biology Commons Hoffmann, Federico G.; Vandewege, Michael W.; Storz, Jay F.; and Opazo, Juan C., "Gene Turnover and Diversification of the α- and β- Globin Gene Families in Sauropsid Vertebrates" (2018). Faculty Publications in the Biological Sciences. 634. https://digitalcommons.unl.edu/bioscifacpub/634 This Article is brought to you for free and open access by the Papers in the Biological Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications in the Biological Sciences by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. GBE Gene Turnover and Diversification of the a-andb-Globin Gene Families in Sauropsid Vertebrates Federico G. Hoffmann1,2,*, Michael W. Vandewege3,JayF.Storz4, and Juan C. Opazo5 1Department of Biochemistry, Molecular Biology, Entomology, and Plant Pathology, Mississippi State University 2Institute for Genomics, Biocomputing and Biotechnology, Mississippi State University 3Department of Biological Sciences, Texas Tech University 4School of Biological Sciences, University of Nebraska 5Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile *Corresponding author: E-mail: [email protected]. -

Germline -Specification, Sex, and Stem Cells-”
CONTENTS Introduction・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 1 Organization of the National Institute for Basic Biology ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 2 Goals of the National Institute for Basic Biology ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 5 Personnel Changes and Awardees in 2012 ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 7 Cell Biology Division of Cell Mechanisms・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 8 Division of Intercellular Signaling Biology・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 11 Laboratory of Neuronal Cell Biology ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 14 Laboratory of Cell Sociology・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 16 Developmental Biology Division of Morphogenesis ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 17 Division of Developmental Genetics ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 20 Division of Molecular and Developmental Biology ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 23 Division of Embryology ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 26 Division of Germ Cell Biology・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・ 29 Laboratory of Molecular Genetics for Reproduction ・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・