Glutaredoxin 1 Protects Dopaminergic Cells by Increased Protein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![Downloaded from [266]](https://docslib.b-cdn.net/cover/7352/downloaded-from-266-347352.webp)
Downloaded from [266]
Patterns of DNA methylation on the human X chromosome and use in analyzing X-chromosome inactivation by Allison Marie Cotton B.Sc., The University of Guelph, 2005 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2012 © Allison Marie Cotton, 2012 Abstract The process of X-chromosome inactivation achieves dosage compensation between mammalian males and females. In females one X chromosome is transcriptionally silenced through a variety of epigenetic modifications including DNA methylation. Most X-linked genes are subject to X-chromosome inactivation and only expressed from the active X chromosome. On the inactive X chromosome, the CpG island promoters of genes subject to X-chromosome inactivation are methylated in their promoter regions, while genes which escape from X- chromosome inactivation have unmethylated CpG island promoters on both the active and inactive X chromosomes. The first objective of this thesis was to determine if the DNA methylation of CpG island promoters could be used to accurately predict X chromosome inactivation status. The second objective was to use DNA methylation to predict X-chromosome inactivation status in a variety of tissues. A comparison of blood, muscle, kidney and neural tissues revealed tissue-specific X-chromosome inactivation, in which 12% of genes escaped from X-chromosome inactivation in some, but not all, tissues. X-linked DNA methylation analysis of placental tissues predicted four times higher escape from X-chromosome inactivation than in any other tissue. Despite the hypomethylation of repetitive elements on both the X chromosome and the autosomes, no changes were detected in the frequency or intensity of placental Cot-1 holes. -
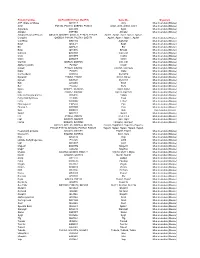
Gene Ids Organism ATP Citrate Synthase Q3V117 Acly
Protein Families UniProtKB ID (from MudPIT) Gene IDs Organism ATP citrate synthase Q3V117 Acly Mus musculus (Mouse) Actin P68134, P60710, Q8BFZ3, P68033 Acta1, Actb, Actbl2, Actc1 Mus musculus (Mouse) Argonaute Q8CJG0 Ago2 Mus musculus (Mouse) Ahnak2 E9PYB0 Ahnak2 Mus musculus (Mouse) Adaptor Related Protein Q8CC13, Q8CBB7, Q3UHJ0, P17426, P17427, Ap1b1, Ap1g1, Aak1, Ap2a1, Ap2a2, Complex Q9DBG3, P84091, P62743, Q9Z1T1 Ap2b1, Ap2m1, Ap2s1 , Ap3b1 Mus musculus (Mouse) V-ATPase Q9Z1G4 Atp6v0a1 Mus musculus (Mouse) Bag3 Q9JLV1 Bag3 Mus musculus (Mouse) Bcr Q6PAJ1 Bcr Mus musculus (Mouse) Bmp Q91Z96 Bmp2k Mus musculus (Mouse) Calcoco Q8CGU1 Calcoco1 Mus musculus (Mouse) Ccdc D3YZP9 Ccdc6 Mus musculus (Mouse) Clint1 Q5SUH7 Clint1 Mus musculus (Mouse) Clathrin Q6IRU5, Q68FD5 Cltb, Cltc Mus musculus (Mouse) Alpha-crystallin P23927 Cryab Mus musculus (Mouse) Casein P19228, Q02862 Csn1s1, Csn1s2a Mus musculus (Mouse) Dab2 P98078 Dab2 Mus musculus (Mouse) Connecdenn Q8K382 Dennd1a Mus musculus (Mouse) Dynamin P39053, P39054 Dnm1, Dnm2 Mus musculus (Mouse) Dynein Q9JHU4 Dync1h1 Mus musculus (Mouse) Edc Q3UJB9 Edc4 Mus musculus (Mouse) Eef P58252 Eef2 Mus musculus (Mouse) Epsin Q80VP1, Q5NCM5 Epn1, Epn2 Mus musculus (Mouse) Eps P42567, Q60902 Eps15, Eps15l1 Mus musculus (Mouse) Fatty acid binding protein Q05816 Fabp5 Mus musculus (Mouse) Fatty Acid Synthase P19096 Fasn Mus musculus (Mouse) Fcho Q3UQN2 Fcho2 Mus musculus (Mouse) Fibrinogen A E9PV24 Fga Mus musculus (Mouse) Filamin A Q8BTM8 Flna Mus musculus (Mouse) Gak Q99KY4 Gak Mus musculus (Mouse) -

Characterization of the Macrophage Transcriptome in Glomerulonephritis-Susceptible and -Resistant Rat Strains
Genes and Immunity (2011) 12, 78–89 & 2011 Macmillan Publishers Limited All rights reserved 1466-4879/11 www.nature.com/gene ORIGINAL ARTICLE Characterization of the macrophage transcriptome in glomerulonephritis-susceptible and -resistant rat strains K Maratou1, J Behmoaras2, C Fewings1, P Srivastava1, Z D’Souza1, J Smith3, L Game4, T Cook2 and T Aitman1 1Physiological Genomics and Medicine Group, MRC Clinical Sciences Centre, Imperial College London, London, UK; 2Centre for Complement and Inflammation Research, Imperial College London, London, UK; 3Renal Section, Imperial College London, London, UK and 4Genomics Laboratory, MRC Clinical Sciences Centre, London, UK Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is unknown. Wistar–Kyoto (WKY) rats show marked susceptibility to CRGN, whereas Lewis rats are resistant. Glomerular injury and crescent formation are macrophage dependent and mainly explained by seven quantitative trait loci (Crgn1–7). Here, we used microarray analysis in basal and lipopolysaccharide (LPS)-stimulated macrophages to identify genes that reside on pathways predisposing WKY rats to CRGN. We detected 97 novel positional candidates for the uncharacterized Crgn3–7. We identified 10 additional secondary effector genes with profound differences in expression between the two strains (45-fold change, o1% false discovery rate) for basal and LPS-stimulated macrophages. Moreover, we identified eight genes with differentially expressed alternatively spliced isoforms, by using an in-depth analysis at the probe level that allowed us to discard false positives owing to polymorphisms between the two rat strains. Pathway analysis identified several common linked pathways, enriched for differentially expressed genes, which affect macrophage activation. -

Interactome of Mirnas and Transcriptome of Human Umbilical Cord Endothelial Cells Exposed to Short-Term Simulated Microgravity
www.nature.com/npjmgrav ARTICLE OPEN Interactome of miRNAs and transcriptome of human umbilical cord endothelial cells exposed to short-term simulated microgravity Dharanibalan Kasiviswanathan1,2,4, Rajadurai Chinnasamy Perumal 3,4, Srinivasan Bhuvaneswari1,2, Pavitra Kumar1, ✉ Lakshmikirupa Sundaresan 1,2, Manuel Philip 3, Sajesh Puthenpurackal Krishnankutty3 and Suvro Chatterjee1,2 Adaptation of humans in low gravity conditions is a matter of utmost importance when efforts are on to a gigantic leap in human space expeditions for tourism and formation of space colonies. In this connection, cardiovascular adaptation in low gravity is a critical component of human space exploration. Deep high-throughput sequencing approach allowed us to analyze the miRNA and mRNA expression profiles in human umbilical cord vein endothelial cells (HUVEC), cultured under gravity (G), and stimulated microgravity (MG) achieved with a clinostat. The present study identified totally 1870 miRNAs differentially expressed in HUVEC under MG condition when compared to the cells subjected to unitary G conditions. The functional association of identified miRNAs targeting specific mRNAs revealed that miRNAs, hsa-mir-496, hsa-mir-151a, hsa-miR-296-3p, hsa-mir-148a, hsa-miR-365b-5p, hsa- miR-3687, hsa-mir-454, hsa-miR-155-5p, and hsa-miR-145-5p differentially regulated the genes involved in cell adhesion, angiogenesis, cell cycle, JAK-STAT signaling, MAPK signaling, nitric oxide signaling, VEGF signaling, and wound healing pathways. Further, the q-PCR based experimental studies of upregulated and downregulated miRNA and mRNAs demonstrate that the above 1234567890():,; reported miRNAs influence the cell proliferation and vascular functions of the HUVEC in MG conditions effectively. -

Identification of a Microdeletion at Xp22.13 in a Taiwanese Family
Journal of Human Genetics (2011) 56, 8–11 & 2011 The Japan Society of Human Genetics All rights reserved 1434-5161/11 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Identification of a microdeletion at Xp22.13 in a Taiwanese family presenting with Nance-Horan syndrome Hsiao-Mei Liao1, Dau-Ming Niu2,3, Yan-Jang Chen4,5, Jye-Siung Fang6, Shih-Jen Chen7 and Chia-Hsiang Chen8,9,10 Nance-Horan syndrome (NHS) is a rare X-linked disorder characterized by congenital cataracts, dental anomalies and mental retardation. The disease has been linked to a novel gene termed NHS located at Xp22.13. The majority of pathogenic mutations of the disease include nonsense mutations and small deletions and insertions that lead to truncation of the NHS protein. In this study, we identified a microdeletion of B0.92 Mb at Xp22.13 detected by array-based comparative genomic hybridization in two brothers presenting congenital cataract, dental anomalies, facial dysmorphisms and mental retardation. The deleted region encompasses the REPS2, NHS, SCML1 and RAI2 genes, and was transmitted from their carrier mother who presented only mild cataract. Our findings are in line with several recent case reports to indicate that genomic rearrangement involving the NHS gene is an important genetic etiology underlying NHS. Journal of Human Genetics (2011) 56, 8–11; doi:10.1038/jhg.2010.121; published online 30 September 2010 Keywords: array CGH; congenital cataract; mental retardation; microdeletion; Nance-Horan syndrome INTRODUCTION associated with patient with NHS or X-linked congenital cataract,6,12 Nance-Horan syndrome (NHS) (OMIM 302350) is a rare X-linked suggesting that genomic rearrangement also contributes to the genetic genetic disorder characterized by congenital cataracts, dental defects, etiology underlying the NHS. -

REPS2 Antibody (N-Term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap13131a
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 REPS2 Antibody (N-term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP13131a Specification REPS2 Antibody (N-term) - Product Information Application WB, IHC-P,E Primary Accession Q8NFH8 Other Accession NP_004717.2, NP_001074444.1 Reactivity Human, Mouse Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 71534 Antigen Region 153-182 REPS2 Antibody (N-term) - Additional Information REPS2 Antibody (N-term) (Cat. #AP13131a) Gene ID 9185 western blot analysis in mouse brain tissue lysates (35ug/lane).This demonstrates the Other Names REPS2 antibody detected the REPS2 protein RalBP1-associated Eps domain-containing (arrow). protein 2, Partner of RalBP1, RalBP1-interacting protein 2, REPS2, POB1 Target/Specificity This REPS2 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 153-182 amino acids from the N-terminal region of human REPS2. Dilution WB~~1:1000 IHC-P~~1:10~50 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. REPS2 Antibody (N-term) (Cat. #AP13131a)immunohistochemistry analysis Storage in formalin fixed and paraffin embedded Maintain refrigerated at 2-8°C for up to 2 human cerebellum tissue followed by weeks. For long term storage store at -20°C peroxidase conjugation of the secondary in small aliquots to prevent freeze-thaw antibody and DAB staining.This data cycles. demonstrates the use of REPS2 Antibody (N-term) for immunohistochemistry. Clinical Precautions relevance has not been evaluated. -

A Temporally Controlled Sequence of X-Chromosome Inactivation and Reactivation Defines Female Mouse in Vitro Germ Cells with Meiotic Potential
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.11.455976; this version posted August 11, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. A temporally controlled sequence of X-chromosome inactivation and reactivation defines female mouse in vitro germ cells with meiotic potential Jacqueline Severino1†, Moritz Bauer1,9†, Tom Mattimoe1, Niccolò Arecco1, Luca Cozzuto1, Patricia Lorden2, Norio Hamada3, Yoshiaki Nosaka4,5,6, So Nagaoka4,5,6, Holger Heyn2, Katsuhiko Hayashi7, Mitinori Saitou4,5,6 and Bernhard Payer1,8* Abstract The early mammalian germ cell lineage is characterized by extensive epigenetic reprogramming, which is required for the maturation into functional eggs and sperm. In particular, the epigenome needs to be reset before parental marks can be established and then transmitted to the next generation. In the female germ line, reactivation of the inactive X- chromosome is one of the most prominent epigenetic reprogramming events, and despite its scale involving an entire chromosome affecting hundreds of genes, very little is known about its kinetics and biological function. Here we investigate X-chromosome inactivation and reactivation dynamics by employing a tailor-made in vitro system to visualize the X-status during differentiation of primordial germ cell-like cells (PGCLCs) from female mouse embryonic stem cells (ESCs). We find that the degree of X-inactivation in PGCLCs is moderate when compared to somatic cells and characterized by a large number of genes escaping full inactivation. -

Zimmer Cell Calcium 2013 Mammalian S100 Evolution.Pdf
Cell Calcium 53 (2013) 170–179 Contents lists available at SciVerse ScienceDirect Cell Calcium jo urnal homepage: www.elsevier.com/locate/ceca Evolution of the S100 family of calcium sensor proteins a,∗ b b,1 b Danna B. Zimmer , Jeannine O. Eubanks , Dhivya Ramakrishnan , Michael F. Criscitiello a Center for Biomolecular Therapeutics and Department of Biochemistry & Molecular Biology, University of Maryland School of Medicine, 108 North Greene Street, Baltimore, MD 20102, United States b Comparative Immunogenetics Laboratory, Department of Veterinary Pathobiology, College of Veterinary Medicine & Biomedical Sciences, Texas A&M University, College Station, TX 77843-4467, United States a r t i c l e i n f o a b s t r a c t 2+ Article history: The S100s are a large group of Ca sensors found exclusively in vertebrates. Transcriptomic and genomic Received 4 October 2012 data from the major radiations of mammals were used to derive the evolution of the mammalian Received in revised form 1 November 2012 S100s genes. In human and mouse, S100s and S100 fused-type proteins are in a separate clade from Accepted 3 November 2012 2+ other Ca sensor proteins, indicating that an ancient bifurcation between these two gene lineages Available online 14 December 2012 has occurred. Furthermore, the five genomic loci containing S100 genes have remained largely intact during the past 165 million years since the shared ancestor of egg-laying and placental mammals. Keywords: Nonetheless, interesting births and deaths of S100 genes have occurred during mammalian evolution. Mammals The S100A7 loci exhibited the most plasticity and phylogenetic analyses clarified relationships between Phylogenetic analyses the S100A7 proteins encoded in the various mammalian genomes. -

Nuclear DICKKOPF-1 As a Biomarker of Chemoresistance and Poor Clinical Outcome in Colorectal Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No.8 Nuclear DICKKOPF-1 as a biomarker of chemoresistance and poor clinical outcome in colorectal cancer Óscar Aguilera1,2,*, José Manuel González-Sancho1,*, Sandra Zazo2, Raúl Rincón2, Agustín F. Fernández3, Olga Tapia4,11, Francesc Canals5, Beatriz Morte1,6, Vincenzo Calvanese3,13, José L. Orgaz1,12, Núria Niell1, Susana Aguilar7, José M. Freije8, Osvaldo Graña9, David G. Pisano9, Aurea Borrero2, Javier Martínez-Useros2, Benilde Jiménez1, Mario F. Fraga3,10, Jesús García-Foncillas2, Carlos López-Otín8, Miguel Lafarga4, Federico Rojo2 and Alberto Muñoz1 1 Instituto de Investigaciones Biomédicas “Alberto Sols”, Consejo Superior de Investigaciones Científicas (CSIC), Universidad Autónoma de Madrid, Madrid, Spain 2 Instituto de Investigación Sanitaria-Fundación Jiménez Díaz, Madrid, Spain 3 Cancer Epigenetics Laboratory, Instituto Universitario de Oncología del Principado de Asturias (IUOPA-HUCA), Universidad de Oviedo, Oviedo, Spain 4 Departamento de Anatomía y Biología Celular, Universidad de Cantabria-IFIMAV, Santander, Spain 5 Proteomics Laboratory, Vall d’Hebron Institute of Oncology (VHIO), Vall d’Hebron University Hospital, Barcelona, Spain 6 Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), Madrid, Spain 7 Catalan Institute of Oncology-IDIBELL, L’Hospitalet de Llobregat, Barcelona, Spain 8 Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain 9 Bioinformatics Unit -

Tepzz 8Z6z54a T
(19) TZZ ZZ_T (11) EP 2 806 054 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 26.11.2014 Bulletin 2014/48 C40B 40/06 (2006.01) C12Q 1/68 (2006.01) C40B 30/04 (2006.01) C07H 21/00 (2006.01) (21) Application number: 14175049.7 (22) Date of filing: 28.05.2009 (84) Designated Contracting States: (74) Representative: Irvine, Jonquil Claire AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HGF Limited HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL 140 London Wall PT RO SE SI SK TR London EC2Y 5DN (GB) (30) Priority: 28.05.2008 US 56827 P Remarks: •Thecomplete document including Reference Tables (62) Document number(s) of the earlier application(s) in and the Sequence Listing can be downloaded from accordance with Art. 76 EPC: the EPO website 09753364.0 / 2 291 553 •This application was filed on 30-06-2014 as a divisional application to the application mentioned (71) Applicant: Genomedx Biosciences Inc. under INID code 62. Vancouver, British Columbia V6J 1J8 (CA) •Claims filed after the date of filing of the application/ after the date of receipt of the divisional application (72) Inventor: Davicioni, Elai R.68(4) EPC). Vancouver British Columbia V6J 1J8 (CA) (54) Systems and methods for expression- based discrimination of distinct clinical disease states in prostate cancer (57) A system for expression-based discrimination of distinct clinical disease states in prostate cancer is provided that is based on the identification of sets of gene transcripts, which are characterized in that changes in expression of each gene transcript within a set of gene transcripts can be correlated with recurrent or non- recur- rent prostate cancer. -

The Ef-Hand Ca2+-Binding Protein Super-Family: a Genome-Wide Analysis of Gene Expression Patterns in the Adult Mouse Brain
View metadata, citation and similar papers at core.ac.uk brought to you by CORE Published in 1HXURVFLHQFH± which should be cited to refer to this work. provided by RERO DOC Digital Library THE EF-HAND CA2+-BINDING PROTEIN SUPER-FAMILY: A GENOME-WIDE ANALYSIS OF GENE EXPRESSION PATTERNS IN THE ADULT MOUSE BRAIN F. GIRARD, * J. VENAIL B. SCHWALLER AND M. R. CELIO ganglia, hippocampus, amygdala, thalamus, hypothalamus, Anatomy and Program in Neuroscience, Department of cerebellum, midbrain, pons and medulla, and on clearly Medicine, Faculty of Science, University of Fribourg, Route A. identifiable sub-structures within each of these areas. The Gockel 1, CH-1700 Fribourg, Switzerland expression profiles of four family-members, namely hippocalcin-like 4, neurocalcin-d, plastin 3 and tescalcin, that have not been hitherto reported, at either the mRNA Abstract—In mice, 249 putative members of the superfamily (in-situ-hybridization) or the protein (immunohistochemical) of EF-hand domain Ca2+-binding proteins, manifesting levels, are now presented for the first time. The fruit of great diversity in structure, cellular localization and our analysis is a document in which the gene-expression functions have been identified. Three members in particular, profiles of all members of the EF-hand family genes namely, calbindin-D28K, calretinin and parvalbumin, are compared, and in which future possible neuronal are widely used as markers for specific neuronal markers for specific cells/brain areas are identified. The subpopulations in different regions of the brain. The aim assembled information could afford functional clues to of the present study was to compile a comprehensive atlas investigators, conducive to further experimental pursuit. -

Detection of H3k4me3 Identifies Neurohiv Signatures, Genomic
viruses Article Detection of H3K4me3 Identifies NeuroHIV Signatures, Genomic Effects of Methamphetamine and Addiction Pathways in Postmortem HIV+ Brain Specimens that Are Not Amenable to Transcriptome Analysis Liana Basova 1, Alexander Lindsey 1, Anne Marie McGovern 1, Ronald J. Ellis 2 and Maria Cecilia Garibaldi Marcondes 1,* 1 San Diego Biomedical Research Institute, San Diego, CA 92121, USA; [email protected] (L.B.); [email protected] (A.L.); [email protected] (A.M.M.) 2 Departments of Neurosciences and Psychiatry, University of California San Diego, San Diego, CA 92103, USA; [email protected] * Correspondence: [email protected] Abstract: Human postmortem specimens are extremely valuable resources for investigating trans- lational hypotheses. Tissue repositories collect clinically assessed specimens from people with and without HIV, including age, viral load, treatments, substance use patterns and cognitive functions. One challenge is the limited number of specimens suitable for transcriptional studies, mainly due to poor RNA quality resulting from long postmortem intervals. We hypothesized that epigenomic Citation: Basova, L.; Lindsey, A.; signatures would be more stable than RNA for assessing global changes associated with outcomes McGovern, A.M.; Ellis, R.J.; of interest. We found that H3K27Ac or RNA Polymerase (Pol) were not consistently detected by Marcondes, M.C.G. Detection of H3K4me3 Identifies NeuroHIV Chromatin Immunoprecipitation (ChIP), while the enhancer H3K4me3 histone modification was Signatures, Genomic Effects of abundant and stable up to the 72 h postmortem. We tested our ability to use H3K4me3 in human Methamphetamine and Addiction prefrontal cortex from HIV+ individuals meeting criteria for methamphetamine use disorder or not Pathways in Postmortem HIV+ Brain (Meth +/−) which exhibited poor RNA quality and were not suitable for transcriptional profiling.