Brachyury Mice
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table S1. Nakharuthai and Srisapoome (2020)
Primer name Nucleotide sequence (5’→3’) Purpose CXC-1F New TGAACCCTGAGCTGAAGGCCGTGA Real-time PCR CXC-1R New TGAAGGTCTGATGAGTTTGTCGTC Real-time PCR CXC-1R CCTTCAGCTCAGGGTTCAAGC Genomic PCR CXC-2F New GCTTGAACCCCGAGCTGAAAAACG Real-time PCR CXC-2R New GTTCAGAGGTCGTATGAGGTGCTT Real-time PCR CXC-2F CAAGCAGGACAACAGTGTCTGTGT 3’RACE CXC-2AR GTTGCATGATTTGGATGCTGGGTAG 5’RACE CXC-1FSB AACATATGTCTCCCAGGCCCAACTCAAAC Southern blot CXC-1RSB CTCGAGTTATTTTGCACTGATGTGCAA Southern blot CXC1Exon1F CAAAGTGTTTCTGCTCCTGG Genomic PCR On-CXC1FOverEx CATATGCAACTCAAACAAGCAGGACAACAGT Overexpression On-CXC1ROverEx CTCGAGTTTTGCACTGATGTGCAATTTCAA Overexpression On-CXC2FOverEx CATATGCAACTCAAACAAGCAGGACAACAGT Overexpression On-CXC2ROverEx CTCGAGCATGGCAGCTGTGGAGGGTTCCAC Overexpression β-actinrealtimeF ACAGGATGCAGAAGGAGATCACAG Real-time PCR β-actinrealtimeR GTACTCCTGCTTGCTGATCCACAT Real-time PCR M13F AAAACGACGGCCAG Nucleotide sequencing M13R AACAGCTATGACCATG Nucleotide sequencing UPM-long (0.4 µM) CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT RACE PCR UPM-short (2 µM) CTAATAC GACTCACTATA GGGC RACE PCR Table S1. Nakharuthai and Srisapoome (2020) On-CXC1 Nucleotide (%) Amino acid (%) On-CXC2 Nucleotide (%) Amino acid (%) Versus identity identity similarity Versus identity identity Similarity Teleost fish 1. Rock bream, Oplegnathus fasciatus (AB703273) 64.5 49.1 68.1 O. fasciatus 70.7 57.7 75.4 2. Mandarin fish, Siniperca chuatsi (AAY79282) 63.2 48.1 68.9 S. chuatsi 70.5 54.0 78.8 3. Atlantic halibut, Hippoglossus hippoglossus (ACY54778) 52.0 39.3 51.1 H. hippoglossus 64.5 46.3 63.9 4. Common carp IL-8, Cyprinus carpio (ABE47600) 44.9 19.1 34.1 C. carpio 49.4 21.9 42.6 5. Rainbow trout IL-8, Oncorhynchus mykiss (CAC33585) 44.0 21.3 36.3 O. mykiss 47.3 23.7 44.4 6. Japanese flounder IL-8, Paralichthys olivaceus (AAL05442) 48.4 25.4 45.9 P. -

UTX Regulates Mesoderm Differentiation of Embryonic Stem Cells Independent of H3K27 Demethylase Activity
UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity Chaochen Wanga, Ji-Eun Leea,1, Young-Wook Chob,1, Ying Xiaoc, Qihuang Jina, Chengyu Liud, and Kai Gea,2 aLaboratory of Endocrinology and Receptor Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892; bKorea Basic Science Institute Chuncheon Center, Chuncheon, Kangwon, Korea 200-701; cDermatology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; and dTransgenic Core, Center for Molecular Medicine, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892 Edited by Mark Groudine, Fred Hutchinson Cancer Research Center, Seattle, WA, and approved August 3, 2012 (received for review March 9, 2012) To investigate the role of histone H3K27 demethylase UTX in male cells. UTY is a paralog of the X-linked UTX and shares embryonic stem (ES) cell differentiation, we have generated UTX 88% sequence homology with UTX protein. Unlike UTX, UTY knockout (KO) and enzyme-dead knock-in male ES cells. Deletion lacks detectable histone demethlase activity in vitro (8, 12). The of the X-chromosome-encoded UTX gene in male ES cells markedly viability data from male and female UTX KO mice indicate a decreases expression of the paralogous UTY gene encoded by Y largely functional redundancy between UTX and UTY during chromosome, but has no effect on global H3K27me3 level, Hox male embryonic development (13). gene expression, or ES cell self-renewal. However, UTX KO cells UTX has been shown to regulate myocyte differentiation, heart show severe defects in mesoderm differentiation and induction of development, and T-box transcription factor target gene expres- Brachyury, a transcription factor essential for mesoderm develop- sion (13–15). -

Mouse Models of Human Disease an Evolutionary Perspective Robert L
170 commentary Evolution, Medicine, and Public Health [2016] pp. 170–176 doi:10.1093/emph/eow014 Mouse models of human disease An evolutionary perspective Robert L. Perlman* Department of Pediatrics, The University of Chicago, 5841 S. Maryland Ave, MC 5058, Chicago, IL 60637, USA *E-mail: [email protected] Received 31 December 2015; revised version accepted 12 April 2016 ABSTRACT The use of mice as model organisms to study human biology is predicated on the genetic and physio- logical similarities between the species. Nonetheless, mice and humans have evolved in and become adapted to different environments and so, despite their phylogenetic relatedness, they have become very different organisms. Mice often respond to experimental interventions in ways that differ strikingly from humans. Mice are invaluable for studying biological processes that have been conserved during the evolution of the rodent and primate lineages and for investigating the developmental mechanisms by which the conserved mammalian genome gives rise to a variety of different species. Mice are less reliable as models of human disease, however, because the networks linking genes to disease are likely to differ between the two species. The use of mice in biomedical research needs to take account of the evolved differences as well as the similarities between mice and humans. KEYWORDS: allometry; cancer; gene networks; life history; model organisms transgenic, knockout, and knockin mice, have If you have cancer and you are a mouse, we can provided added impetus and powerful tools for take good care of you. Judah Folkman [1] mouse research, and have led to a dramatic increase in the use of mice as model organisms. -

Micromys Minutus)
Acta Theriol (2013) 58:101–104 DOI 10.1007/s13364-012-0102-0 SHORT COMMUNICATION The origin of Swedish and Norwegian populations of the Eurasian harvest mouse (Micromys minutus) Lars Råberg & Jon Loman & Olof Hellgren & Jeroen van der Kooij & Kjell Isaksen & Roar Solheim Received: 7 May 2012 /Accepted: 17 September 2012 /Published online: 29 September 2012 # Mammal Research Institute, Polish Academy of Sciences, Białowieża, Poland 2012 Abstract The harvest mouse (Micromys minutus) occurs the Mediterranean, from France to Russia and northwards throughout most of continental Europe. There are also two to central Finland (Mitchell-Jones et al. 1999). Recently, isolated and recently discovered populations on the it has also been found to occur in Sweden and Norway. Scandinavian peninsula, in Sweden and Norway. Here, we In 1985, an isolated population was discovered in the investigate the origin of these populations through analyses province of Dalsland in western Sweden (Loman 1986), of mitochondrial DNA. We found that the two populations and during the last decade, this population has been on the Scandinavian peninsula have different mtDNA found to extend into the surrounding provinces as well haplotypes. A comparison of our haplotypes to published as into Norway. The known distribution in Norway is sequences from most of Europe showed that all Swedish and limited to a relatively small area close to the Swedish Norwegian haplotypes are most closely related to the border, in Eidskog, Hedmark (van der Kooij et al. 2001; haplotypes in harvest mice from Denmark. Hence, the two van der Kooij et al., unpublished data). In 2007, another populations seem to represent independent colonisations but population was discovered in the province of Skåne in originate from the same geographical area. -

Speaker Abstracts 2018 International Chordoma Research Workshop | Speaker Abstracts 1
Sixth International Chordoma Research Workshop Speaker abstracts 2018 International Chordoma Research Workshop | Speaker abstracts 1 TABLE OF CONTENTS EPIGENETIC CONTROL OF BRACHYURY AND METABOLIC STRESS RESPONSE: NOVEL THERAPEUTIC TARGETS FOR CHORDOMA ........................................................................................................................................................... 2 SYSTEMATIC DISCOVERY OF NOVEL VULNERABILITIES IN CHORDOMA .................................................................. 3 RATIONALE FOR THE ADVANCEMENT OF PTEN/AKT PATHWAY INHIBITORS AND COMBINATIONS FOR PERSONALIZED CHORDOMA THERAPY ................................................................................................................... 4 THE ROLE OF PHILANTHROPY IN TRANSFORMING CANCER RESEARCH .................................................................. 5 BRACHYURY IN CHORDOMA AND CARCINOMAS: BIOLOGY AND POTENTIAL TARGETING APPROACHES ............... 6 CHARTING BRACHYURY-MEDIATED DEVELOPMENTAL PATHWAYS DURING EARLY MOUSE EMBRYOGENESIS ...... 7 CRYSTAL STRUCTURES OF BRACHYURY: A PRELUDE TO DRUG DISCOVERY ............................................................ 8 A BRACHYURY TRANSCRIPTIONAL REPORTER TO GUIDE DRUG DISCOVERY........................................................... 9 INVESTIGATING BRACHYURY GENE REGULATION TO IDENTIFY THERAPEUTIC TARGETS IN CHORDOMA ............. 10 DELINEATING AND TARGETING THE BRACHYURY-YAP REGULATORY AXIS IN CANCER ......................................... 11 OPEN ACCESS -

Bangor University DOCTOR of PHILOSOPHY Brachyury in the Human Colon and Colorectal Cancer Williams, Jason
Bangor University DOCTOR OF PHILOSOPHY Brachyury in the Human Colon and Colorectal Cancer Williams, Jason Award date: 2018 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 04. Oct. 2021 Brachyury in the Human Colon and Colorectal Cancer Ph. D. Thesis 2017 Jason Saunders Williams i Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: Title: Supervisor/Department: Funding body (if any): Qualification/Degree obtained: PhD This item is a product of my own research endeavours and is covered by the agreement below in which the item is referred to as “the Work”. It is identical in content to that deposited in the Library, subject to point 4 below. -

A Guide to Mites
A GUIDE TO MITES concentrated in areas where clothes constrict the body, or MITES in areas like the armpits or under the breasts. These bites Mites are arachnids, belonging to the same group as can be extremely itchy and may cause emotional distress. ticks and spiders. Adult mites have eight legs and are Scratching the affected area may lead to secondary very small—sometimes microscopic—in size. They are bacterial infections. Rat and bird mites are very small, a very diverse group of arthropods that can be found in approximately the size of the period at the end of this just about any habitat. Mites are scavengers, predators, sentence. They are quite active and will enter the living or parasites of plants, insects and animals. Some mites areas of a home when their hosts (rats or birds) have left can transmit diseases, cause agricultural losses, affect or have died. Heavy infestations may cause some mites honeybee colonies, or cause dermatitis and allergies in to search for additional blood meals. Unfed females may humans. Although mites such as mold mites go unnoticed live ten days or more after rats have been eliminated. In and have no direct effect on humans, they can become a this area, tropical rat mites are normally associated with nuisance due to their large numbers. Other mites known the roof rat (Rattus rattus), but are also occasionally found to cause a red itchy rash (known as contact dermatitis) on the Norway rat, (R. norvegicus) and house mouse (Mus include a variety of grain and mold mites. Some species musculus). -

A Phylogeographic Survey of the Pygmy Mouse Mus Minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome B Sequences of Museum Specimens
A Phylogeographic Survey of the Pygmy Mouse Mus minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome b Sequences of Museum Specimens Pascale Chevret1*, Terence J. Robinson2, Julie Perez3, Fre´de´ric Veyrunes3, Janice Britton-Davidian3 1 Laboratoire de Biome´trie et Biologie Evolutive, UMR CNRS 5558, Universite´ Lyon 1, Villeurbanne, France, 2 Evolutionary Genomics Group, Department of Botany and Zoology, University of Stellenbosch, Stellenbosch, South Africa, 3 Institut des Sciences de l’Evolution de Montpellier, UMR CNRS 5554, Universite´ Montpellier 2, Montpellier, France Abstract The African pygmy mice (Mus, subgenus Nannomys) are a group of small-sized rodents that occur widely throughout sub- Saharan Africa. Chromosomal diversity within this group is extensive and numerous studies have shown the karyotype to be a useful taxonomic marker. This is pertinent to Mus minutoides populations in South Africa where two different cytotypes (2n = 34, 2n = 18) and a modification of the sex determination system (due to the presence of a Y chromosome in some females) have been recorded. This chromosomal diversity is mirrored by mitochondrial DNA sequences that unambiguously discriminate among the various pygmy mouse species and, importantly, the different M. minutoides cytotypes. However, the geographic delimitation and taxonomy of pygmy mice populations in South Africa is poorly understood. To address this, tissue samples of M. minutoides were taken and analysed from specimens housed in six South African museum collections. Partial cytochrome b sequences (400 pb) were successfully amplified from 44% of the 154 samples processed. Two species were identified: M. indutus and M. minutoides. The sequences of the M. indutus samples provided two unexpected features: i) nuclear copies of the cytochrome b gene were detected in many specimens, and ii) the range of this species was found to extend considerably further south than is presently understood. -

Rats and Mice Have Always Posed a Threat to Human Health
Rats and mice have always posed a threat to human health. Not only do they spread disease but they also cause serious damage to human food and animal feed as well as to buildings, insulation material and electricity cabling. Rats and mice - unwanted house guests! RATS AND MICE ARE AGILE MAMMALS. A mouse can get through a small, 6-7 mm hole (about the diameter of a normal-sized pen) and a rat can get through a 20 mm hole. They can also jump several decimetres at a time. They have no problem climbing up the inside of a vertical sewage pipe and can fall several metres without injuring themselves. Rats are also good swimmers and can be underwater for 5 minutes. IN SWEDEN THERE ARE BASICALLY four different types of rodent that affect us as humans and our housing: the brown rat, the house mouse and the small and large field mouse. THE BROWN RAT (RATTUS NORVEGICUS) THRIVES in all human environments, and especially in damp environments like cellars and sewers. The brown rat is between 20-30 cm in length not counting its tail, which is about 15-23 cm long. These rats normally have brown backs and grey underbellies, but there are also darker ones. They are primarily nocturnal, often keep together in large family groups and dig and gnaw out extensive tunnel systems. Inside these systems, they build large chambers where they store food and build their nests. A pair of rats can produce between 800 and 1000 offspring a year. Since their young are sexually mature and can have offspring of their own at just 2-4 months old, rats reproduce extraordinarily quickly. -

Onecut Transcription Factor OC2 Is a Direct Target of T-Bet in Type-1 T-Helper Cells
Genes and Immunity (2008) 9, 302–308 & 2008 Nature Publishing Group All rights reserved 1466-4879/08 $30.00 www.nature.com/gene ORIGINAL ARTICLE Onecut transcription factor OC2 is a direct target of T-bet in type-1 T-helper cells K Furuno1,2, K Ikeda2, S Hamano3, K Fukuyama1, M Sonoda1, T Hara2, T Sasazuki4 and K Yamamoto1 1Department of Molecular Genetics, Medical Institute of Bioregulation, Kyushu University, Fukuoka, Japan; 2Department of Pediatrics, Graduate School of Medical Science, Kyushu University, Fukuoka, Japan; 3Department of Parasitology, Graduate School of Medical Science, Kyushu University, Fukuoka, Japan and 4International Medical Center of Japan, Tokyo, Japan T-box transcription factor, T-bet, has a central role in the differentiation of T-helper (Th) progenitor cells to Th1 or Th2 effector cells, partly by regulating the expression of genes such as interferon-g (IFN-g). However, the direct target genes, especially those mediating the transcriptional network initiated by T-bet, are not yet fully understood. By combining chromatin immunoprecipitation from Th1 cells with human cytosine-phosphate-guanine-island array analysis, Onecut 2 (OC2), which encodes a member of the ONECUT class of transcriptional activators, was identified as a direct target gene of T-bet. OC2 is expressed in Th1 but not Th2 cells and reporter assays showed that T-bet transactivates OC2 transcription through putative T-bet half-sites locating À451 to À347 of OC2 promoter region. Moreover, we found that OC2 binds and transactivates human T-bet promoter. These results suggest that not only cell-extrinsic regulation via the IFN-g/STAT1 pathway, but also cell-intrinsic transcriptional positive feedback loop between T-bet and OC2 could be involved in Th1 development. -

Mice Are Here to Stay
Mice A re Here to Stay Wherever people live, there are mice. It would be difficult to find another animal that has adapted to the habitats created by humans as well as the house mouse has. It thus seemed obvious to Diethard Tautz at the Max Planck Institute for Evolutionary Biology in Plön that the species would make an ideal model system for investigating how evolution works. BIOLOGY & MEDICINE_Evolution Mice A re Here to Stay TEXT CORNELIA STOLZE he mice at the Max Planck mice themselves only within a range of can be a key factor in the emergence of Institute in Plön live in their 30 to 50 centimeters, they convey high- new species. For Tautz, the house very own house: they have ly complex messages. mouse is a model for the processes of 16 rooms where they can Diethard Tautz and his colleagues evolution: it would be difficult to find form their family clans and have discovered that house mice be- another animal species that lends itself T territories as they see fit. The experi- have naturally only when they live in so well to the study of the genetic ments Tautz and his colleagues carry a familiar environment and interact mechanisms of evolution. out to study such facets as the rodents’ with other members of their species. “Not only is this species extremely communication, behavior and partner- When animals living in the wild are adaptable, as demonstrated by its dis- ships sometimes take months. During captured, they lose their familiar envi- persal all over the globe, but we also this time, the mice are largely left to ronment: everything smells and tastes know its genome better than that of al- their own devices. -
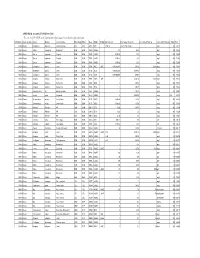
APPENDIX K: Accepted ECOTOX Data Table
APPENDIX K: Accepted ECOTOX Data Table The code list for ECOTOX can be found at: http://cfpub.epa.gov/ecotox/blackbox/help/codelist.pdf CAS Number Chemical Name Genus Species Common Name Effect Group Effect Meas Endpt1 Endpt2 Dur Preferred Conc Value1 Preferred Conc Value2 Preferred Conc Units Preferred % Purity Ref # 330541 Diuron Pimephales promelas Fathead minnow ACC ACC GACC BCF 1 TO 24 0.00315 TO 0.0304 mg/L 100 12612 330541 Diuron Rattus norvegicus Norway rat BCM BCM PHPH NOAEL 140 2500 ppm 100 90555 330541 Diuron Zostera capricorni Eelgrass BCM BCM FLRS LOAEL 8.33E-02 0.01 mg/L 100 72996 330541 Diuron Zostera capricorni Eelgrass BCM BCM FLRS LOAEL 8.33E-02 0.01 mg/L 100 72996 330541 Diuron Zostera capricorni Eelgrass BCM BCM FLRS LOAEL 8.33E-02 0.01 mg/L 100 72996 330541 Diuron Seriatopora hystrix Coral BCM BCM FLRS NOEC LOEC 0.416666667 0.0003 0.0003 mg/L 100 75334 330541 Diuron Seriatopora hystrix Coral BCM BCM FLRS EC25 0.416666667 0.00085 mg/L 100 75334 330541 Diuron Seriatopora hystrix Coral BCM BCM FLRS EC50 0.416666667 0.0023 mg/L 100 75334 330541 Diuron Acropora formosa Stony coral BCM BCM FLRS NOEC LOEC 1 0.00003 0.0003 mg/L 100 75334 330541 Diuron Acropora formosa Stony coral BCM BCM FLRS EC25 1 0.0012 mg/L 100 75334 330541 Diuron Acropora formosa Stony coral BCM BCM FLRS EC50 1 0.0027 mg/L 100 75334 330541 Diuron Synechococcus sp. Blue-green algae BCM BCM CHCT NOAEL 3 0.0033 mg/L 99 98904 330541 Diuron Lemna minor Duckweed BCM BCM GLTH LOAEL 2 0.02475 mg/L >99 64164 330541 Diuron Scenedesmus acutus Green Algae BCM BCM