Structural Basis of the Recognition of the SAMP Motif of Adenomatous Polyposis Coli by the Src-Homology 3 Domain†,‡
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma
Anatomy and Pathology Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma Sarah L. Lake,1 Sarah E. Coupland,1 Azzam F. G. Taktak,2 and Bertil E. Damato3 PURPOSE. To detect deletions and loss of heterozygosity of disease is fatal in 92% of patients within 2 years of diagnosis. chromosome 3 in a rare subset of fatal, disomy 3 uveal mela- Clinical and histopathologic risk factors for UM metastasis noma (UM), undetectable by fluorescence in situ hybridization include large basal tumor diameter (LBD), ciliary body involve- (FISH). ment, epithelioid cytomorphology, extracellular matrix peri- ϩ ETHODS odic acid-Schiff-positive (PAS ) loops, and high mitotic M . Multiplex ligation-dependent probe amplification 3,4 5 (MLPA) with the P027 UM assay was performed on formalin- count. Prescher et al. showed that a nonrandom genetic fixed, paraffin-embedded (FFPE) whole tumor sections from 19 change, monosomy 3, correlates strongly with metastatic death, and the correlation has since been confirmed by several disomy 3 metastasizing UMs. Whole-genome microarray analy- 3,6–10 ses using a single-nucleotide polymorphism microarray (aSNP) groups. Consequently, fluorescence in situ hybridization were performed on frozen tissue samples from four fatal dis- (FISH) detection of chromosome 3 using a centromeric probe omy 3 metastasizing UMs and three disomy 3 tumors with Ͼ5 became routine practice for UM prognostication; however, 5% years’ metastasis-free survival. to 20% of disomy 3 UM patients unexpectedly develop metas- tases.11 Attempts have therefore been made to identify the RESULTS. Two metastasizing UMs that had been classified as minimal region(s) of deletion on chromosome 3.12–15 Despite disomy 3 by FISH analysis of a small tumor sample were found these studies, little progress has been made in defining the key on MLPA analysis to show monosomy 3. -

DDEF1 Monoclonal Antibody (M01), Clone 2G7
Produktinformation Diagnostik & molekulare Diagnostik Laborgeräte & Service Zellkultur & Verbrauchsmaterial Forschungsprodukte & Biochemikalien Weitere Information auf den folgenden Seiten! See the following pages for more information! Lieferung & Zahlungsart Lieferung: frei Haus Bestellung auf Rechnung SZABO-SCANDIC Lieferung: € 10,- HandelsgmbH & Co KG Erstbestellung Vorauskassa Quellenstraße 110, A-1100 Wien T. +43(0)1 489 3961-0 Zuschläge F. +43(0)1 489 3961-7 [email protected] • Mindermengenzuschlag www.szabo-scandic.com • Trockeneiszuschlag • Gefahrgutzuschlag linkedin.com/company/szaboscandic • Expressversand facebook.com/szaboscandic DDEF1 monoclonal antibody (M01), clone 2G7 Catalog # : H00050807-M01 規格 : [ 100 ug ] List All Specification Application Image Product Mouse monoclonal antibody raised against a partial recombinant Western Blot (Cell lysate) Description: DDEF1. Immunogen: DDEF1 (NP_060952, 1030 a.a. ~ 1129 a.a) partial recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Sequence: VQSRDAIQKQASEDSNDLTPTLPETPVPLPRKINTGKNKVRRVKTIYDCQ ADNDDELTFIEGEVIIVTGEEDQEWWIGHIEGQPERKGVFPVSFVHILSD enlarge Western Blot (Cell lysate) Host: Mouse Reactivity: Human, Rat Isotype: IgG2a Kappa Quality Control Antibody Reactive Against Recombinant Protein. enlarge Testing: Western Blot (Recombinant protein) Immunofluorescence enlarge Sandwich ELISA (Recombinant Western Blot detection against Immunogen (36.74 KDa) . protein) Storage Buffer: In 1x PBS, pH 7.4 Storage Store at -20°C or lower. Aliquot to avoid repeated freezing -

Integrative Analysis of Genomic Amplification-Dependent Expression
Oncogene (2020) 39:4118–4131 https://doi.org/10.1038/s41388-020-1279-3 ARTICLE Integrative analysis of genomic amplification-dependent expression and loss-of-function screen identifies ASAP1 as a driver gene in triple-negative breast cancer progression 1 1 1 2 2 Jichao He ● Ronan P. McLaughlin ● Lambert van der Beek ● Sander Canisius ● Lodewyk Wessels ● 3 3 3 1 1 Marcel Smid ● John W. M. Martens ● John A. Foekens ● Yinghui Zhang ● Bob van de Water Received: 26 September 2019 / Revised: 14 March 2020 / Accepted: 17 March 2020 / Published online: 31 March 2020 © The Author(s) 2020. This article is published with open access Abstract The genetically heterogeneous triple-negative breast cancer (TNBC) continues to be an intractable disease, due to lack of effective targeted therapies. Gene amplification is a major event in tumorigenesis. Genes with amplification-dependent expression are being explored as therapeutic targets for cancer treatment. In this study, we have applied Analytical Multi- scale Identification of Recurring Events analysis and transcript quantification in the TNBC genome across 222 TNBC tumors and identified 138 candidate genes with positive correlation in copy number gain (CNG) and gene expression. siRNA-based 1234567890();,: 1234567890();,: loss-of-function screen of the candidate genes has validated EGFR, MYC, ASAP1, IRF2BP2, and CCT5 genes as drivers promoting proliferation in different TNBC cells. MYC, ASAP1, IRF2BP2, and CCT5 display frequent CNG and concurrent expression over 2173 breast cancer tumors (cBioPortal dataset). More frequently are MYC and ASAP1 amplified in TNBC tumors (>30%, n = 320). In particular, high expression of ASAP1, the ADP-ribosylation factor GTPase-activating protein, is significantly related to poor metastatic relapse-free survival of TNBC patients (n = 257, bc-GenExMiner). -
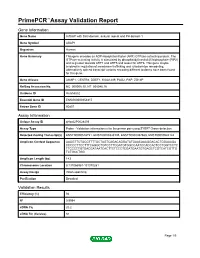
Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 Gene Symbol ASAP1 Organism Human Gene Summary This gene encodes an ADP-ribosylation factor (ARF) GTPase-activating protein. The GTPase-activating activity is stimulated by phosphatidylinositol 45-biphosphate (PIP2) and is greater towards ARF1 and ARF5 and lesser for ARF6. This gene maybe involved in regulation of membrane trafficking and cytoskeleton remodeling. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. Gene Aliases AMAP1, CENTB4, DDEF1, KIAA1249, PAG2, PAP, ZG14P RefSeq Accession No. NC_000008.10, NT_008046.16 UniGene ID Hs.655552 Ensembl Gene ID ENSG00000153317 Entrez Gene ID 50807 Assay Information Unique Assay ID qHsaCIP0028499 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000518721, ENST00000343135, ENST00000357668, ENST00000524124 Amplicon Context Sequence AAGGTTCTGCGTTTTGCTAGTCAGACAGGATATGAACAAAGGACACTGGAAAGA CCCCCTTCCTTTCAGGCTGTCCTTCGATGTGGCCAATCCACCACTCCTGGTCCTC TTCCCCTGTGACGATAATCACTTCTCCCTCGATGAATGTGAGCTCGTCATCGTTG TCTGCCTGG Amplicon Length (bp) 143 Chromosome Location 8:131066961-131070281 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 94 R2 0.9994 cDNA Cq 20.2 cDNA Tm (Celsius) 84 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 38.05 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

Comparative Analyses of Gene Copy Number and Mrna Expression in GBM Tumors And
Title page Comparative analyses of gene copy number and mRNA expression in GBM tumors and GBM xenografts J. Graeme Hodgson # *, Ru-Fang Yeh #, Amrita Ray, Nicholas J Wang, Ivan Smirnov, Mamie Yu, Sujatmi Hariono, Joachim Silber, Heidi S. Feiler, Joe W. Gray, Paul T. Spellman, Scott R. Vandenberg, Mitchel S. Berger, C. David James Departments of Neurological Surgery (JGH, IS, MY, SH, JS, SRV, MSB, CDJ), Pathology (SRV), and Epidemiology and Biostatistics (RY) University of California, San Francisco, CA 94143. Life Sciences Division, Lawrence Berkeley National Lab, Berkeley, CA 94720 (AR, NJW, HSF, JWG, PTS) # These authors contributed equally to this work Running title: Genomic analyses of a GBM xenograft tumor panel * Corresponding Author J. Graeme Hodgson Dept. Neurological Surgery UC San Francisco San Francisco, CA 94143-0808 Phone: 415-476-3630 Fax: 415-476-8218 e-mail: [email protected] Abstract Development of model systems that recapitulate the molecular heterogeneity observed amongst GBM tumors will expedite the testing of targeted molecular therapeutic strategies for GBM treatment. In this study, we profiled DNA copy number and mRNA expression in 21 independent GBM tumor lines maintained as subcutaneous xenografts (GBMX), and compared GBMX molecular signatures to those observed in GBM clinical specimens derived from The Cancer Genome Atlas (TCGA). The predominant copy number signature in both tumor groups was defined by chromosome-7-gain/chromosome-10-loss, a poor prognosis genetic signature. We also observed, at frequencies similar to that detected in TCGA GBMs genomic amplification and overexpression of known GBM oncogenes such as EGFR, MDM2, CDK6 and MYCN, and novel genes including NUP107, SLC35E3, MMP1, MMP13 and DDX1. -

Supplementary Tables S1-S3
Supplementary Table S1: Real time RT-PCR primers COX-2 Forward 5’- CCACTTCAAGGGAGTCTGGA -3’ Reverse 5’- AAGGGCCCTGGTGTAGTAGG -3’ Wnt5a Forward 5’- TGAATAACCCTGTTCAGATGTCA -3’ Reverse 5’- TGTACTGCATGTGGTCCTGA -3’ Spp1 Forward 5'- GACCCATCTCAGAAGCAGAA -3' Reverse 5'- TTCGTCAGATTCATCCGAGT -3' CUGBP2 Forward 5’- ATGCAACAGCTCAACACTGC -3’ Reverse 5’- CAGCGTTGCCAGATTCTGTA -3’ Supplementary Table S2: Genes synergistically regulated by oncogenic Ras and TGF-β AU-rich probe_id Gene Name Gene Symbol element Fold change RasV12 + TGF-β RasV12 TGF-β 1368519_at serine (or cysteine) peptidase inhibitor, clade E, member 1 Serpine1 ARE 42.22 5.53 75.28 1373000_at sushi-repeat-containing protein, X-linked 2 (predicted) Srpx2 19.24 25.59 73.63 1383486_at Transcribed locus --- ARE 5.93 27.94 52.85 1367581_a_at secreted phosphoprotein 1 Spp1 2.46 19.28 49.76 1368359_a_at VGF nerve growth factor inducible Vgf 3.11 4.61 48.10 1392618_at Transcribed locus --- ARE 3.48 24.30 45.76 1398302_at prolactin-like protein F Prlpf ARE 1.39 3.29 45.23 1392264_s_at serine (or cysteine) peptidase inhibitor, clade E, member 1 Serpine1 ARE 24.92 3.67 40.09 1391022_at laminin, beta 3 Lamb3 2.13 3.31 38.15 1384605_at Transcribed locus --- 2.94 14.57 37.91 1367973_at chemokine (C-C motif) ligand 2 Ccl2 ARE 5.47 17.28 37.90 1369249_at progressive ankylosis homolog (mouse) Ank ARE 3.12 8.33 33.58 1398479_at ryanodine receptor 3 Ryr3 ARE 1.42 9.28 29.65 1371194_at tumor necrosis factor alpha induced protein 6 Tnfaip6 ARE 2.95 7.90 29.24 1386344_at Progressive ankylosis homolog (mouse) -

Pathway Entry Into the T Lymphocyte Developmental Molecular Dissection of Prethymic Progenitor
The Journal of Immunology Molecular Dissection of Prethymic Progenitor Entry into the T Lymphocyte Developmental Pathway1 C. Chace Tydell,2 Elizabeth-Sharon David-Fung,2,3 Jonathan E. Moore, Lee Rowen,4 Tom Taghon,5 and Ellen V. Rothenberg6 Notch signaling activates T lineage differentiation from hemopoietic progenitors, but relatively few regulators that initiate this program have been identified, e.g., GATA3 and T cell factor-1 (TCF-1) (gene name Tcf7). To identify additional regulators of T cell specification, a cDNA library from mouse Pro-T cells was screened for genes that are specifically up-regulated in intrathymic T cell precursors as compared with myeloid progenitors. Over 90 genes of interest were iden- tified, and 35 of 44 tested were confirmed to be more highly expressed in T lineage precursors relative to precursors of B and/or myeloid lineage. To a remarkable extent, however, expression of these T lineage-enriched genes, including zinc finger transcription factor, helicase, and signaling adaptor genes, was also shared by stem cells (Lin؊Sca-1؉Kit؉CD27؊) and multipotent progenitors (Lin؊Sca-1؉Kit؉CD27؉), although down-regulated in other lineages. Thus, a major fraction of these early T lineage genes are a regulatory legacy from stem cells. The few genes sharply up-regulated between multipotent progenitors and Pro-T cell stages included those encoding transcription factors Bcl11b, TCF-1 (Tcf7), and HEBalt, Notch target Deltex1, Deltex3L, Fkbp5, Eva1, and Tmem131. Like GATA3 and Deltex1, Bcl11b, Fkbp5, and Eva1 were dependent on Notch/Delta signaling for induction in fetal liver precursors, but only Bcl11b and HEBalt were up-regulated between the first two stages of intrathymic T cell development (double negative 1 and double negative 2) corresponding to T lineage specification. -

Karyotype Aberrations and Gene Mutations Characteristics Correlated It with UM Prognosis Ying Mao1, Bin Li*1 1Beijing Institute of Ophthalmology
New Frontiers in Ophthalmology Review Article ISSN: 2397-2092 Karyotype aberrations and gene mutations characteristics correlated it with UM prognosis Ying Mao1, Bin Li*1 1Beijing Institute of Ophthalmology. Beijing Tongren Eye Center, Beijing Tongren Hospital, Capital Medical University, Beijing Key Laboratory of Ophthalmology and Visual Sciences, Beijing 100005, China Abstract Researchers tried to determined it whether the prognostic of the high incidence karyotype aberrations and gene mutations that had been displayed in uveal melanoma patients. Such as what the prognosis usually was about UM patients with loss of chromosome 3 (monosomy 3), polyploidy (>2N) or gain of chromosome 8q that were combined with/without the main gene mutations of BAP1 (BRCA-associated protein 1), EIF1AX (eukaryotic translation factor 1A), GNAQ (Guanine nucleotide- binding protein, q polypeptide), GNA11 (Guanine nucleotide-binding protein, subunit alpha-11) and SF3B1 (splicing factor 3 subunit B1). Through the literatures of recent years, researchers had suggested aberration of karyotypes and mutation of genes were both made in patients with different prognostic results than we had anticipated but allowed us to reassure patients with a good prognosis. Introduction considered patients’ age, tumor location/size, local tumor invasion through the sclera and epithelioid cell types were not accuracy factors Uveal melanoma (UM) was a most prevalence malignancy tumor for making individualized clinical decisions, so they presented that located in the uveal of the eyes in adults which approximately 50% detected chromosomal alterations were more precise ways to predict of patients had metastatic hepatic disease within 2 to 15 years after the patients’ prognosis than the clinical and pathologic features [9]. -

Table S3: Analysis of Genes Induced in BAHD1 Knockdown HEK213 Cells
Table S3: Analysis of genes induced in BAHD1 knockdown HEK213 cells. Top networks associated to genes induced in BAHD1 KD cells (Ingenuity Pathways Analysis). Associated Network Functions Molecules in Network Score Focus (Molecules in BAHD1-KD transcriptome in upper case) Molecules Cancer, Cell Cycle, Cellular Growth and Proliferation ALCAM, BCL2L11, Caspase, CDKN1C, CLEC11A, CTH, Cyclin 54 27 A, Cyclin E, DACH1, DSP, ERK, GAL, IGF2, Igfbp, IGFBP7, IL6R, KISS1R, LGALS3, Mmp, NEFM, PDGF BB, PHLDA1, Rb, RBL1, RET, RUNX2, SCARA3, SEPP1, SLC6A6, SOX8, SULF1, TAC1, TACSTD1, VCAN, WWP1 Immune Response, Respiratory Disease, Organismal Injury and ADCYAP1R1, ALOX5, Ap1, ATF1, CaMKII, COL13A1, Creb, 39 21 Abnormalities F12, F2R, FSH, hCG, IL1, INDO, ITCH, LDL, MEOX1, NFkB, NID1, NR2F2, P38 MAPK, Pka, Pkc(s), PLA2G7, PLAU, PLC, PLCD3, PRKAR1A, RAP2B, RIPK2, S1PR3, SUMO1, TANK, Tgf beta, TIMP2, Vegf Top Molecular and Cellular Functions associated to genes induced in BAHD1 KD cells (Ingenuity Pathways Analysis). Category Molecules p-value # Molecules Cell Death SAT1, IL6R, PRKAR1A, PHLDA2, RASL10A, PHLDA1, S1PR3, 1,56E-04 - 1,90E-02 43 DSP, IGF2, ADCYAP1R1, ALOX5, QKI, LGALS3, RIPK2, IGFBP7, RUNX2, CDKN1C, PLA2G7, STEAP3, F2R, PLAU, SULF1, KAT2B, RET, BCL2L11, CTH, ATF1, RBL1, DACH1, RTN1, GAL, VCAN, PACS2, TAC1, SUMO1, HLF, EMP2, CLEC11A, TANK, ACSL4, TIMP2, INDO Cellular Growth and Proliferation F12, SAT1, IL6R, PRKAR1A, SEC14L2, PHLDA2, RASL10A, 2,35E-04 - 1,90E-02 46 PHLDA1, DSP, S1PR3, IGF2, CRLF1, ADCYAP1R1, ALOX5, LAMB1, LGALS3, RIPK2, IGFBP7, -

Exon-Level Expression Profiling and Alternative Splicing in Autism Using Lymphoblastoid Cell Lines Zohreh Talebizadeha, Richard Aldenderfera and Xue Wen Chenb
Original article 1 A proof-of-concept study: exon-level expression profiling and alternative splicing in autism using lymphoblastoid cell lines Zohreh Talebizadeha, Richard Aldenderfera and Xue Wen Chenb Objective Autism is a complex, heterogeneous Conclusion The paucity of autism brain samples and neurobehavioral disorder with many causes and varying extensive phenotypic heterogeneity of autism demands degrees of severity. Some genetic implications related finding ways to also identify autism-related genomic events to autism may involve gene-regulatory processes such in accessible nonbrain resources, which may contribute as alternative splicing. Here, we assess the feasibility in biomarker identifications. This proof-of-concept study of profiling exon-level gene expression in autism using shows that the analysis of alternative splicing in the Affymetrix Human exon 1.0 ST array. lymphoblastoid cell line samples has a potential to reveal at least a subset of brain-related deregulation of splicing Methods We examined lymphoblastoid cell line-derived machinery that might be implicated in autism. Psychiatr RNAs from five patients with autism compared with five Genet 24:1–9 c 2014 Wolters Kluwer Health | Lippincott controls. Williams & Wilkins. Results Analysis of variance and Bonferroni Psychiatric Genetics 2014, 24:1–9 multiple test correction identified 57 genes exhibiting differential exon-level expression, suggesting Keywords: alternative splicing, autism, differential expression, exon array, lymphoblastoid cell lines potential changes in the resultant alternatively spliced transcripts in autism compared with controls. Genes aSection of Medical Genetics and Molecular Medicine, Children’s Mercy Hospital and University of Missouri–Kansas City School of Medicine, Kansas City, with differentially expressed exons included CYFIP1, Missouri and bDepartment of Electrical Engineering and Computer Science, a previously reported autism susceptibility gene. -

Genomic and Epigenomic Profile of Uterine Smooth Muscle
International Journal of Molecular Sciences Article Genomic and Epigenomic Profile of Uterine Smooth Muscle Tumors of Uncertain Malignant Potential (STUMPs) Revealed Similarities and Differences with Leiomyomas and Leiomyosarcomas Donatella Conconi 1,*,† , Serena Redaelli 1,† , Andrea Alberto Lissoni 1,2, Chiara Cilibrasi 3, Patrizia Perego 4, Eugenio Gautiero 5, Elena Sala 5, Mariachiara Paderno 1,2, Leda Dalprà 1, Fabio Landoni 1,2, Marialuisa Lavitrano 1 , Gaia Roversi 1,5 and Angela Bentivegna 1,* 1 School of Medicine and Surgery, University of Milano-Bicocca, 20900 Monza, Italy; [email protected] (S.R.); [email protected] (A.A.L.); [email protected] (M.P.); [email protected] (L.D.); [email protected] (F.L.); [email protected] (M.L.); [email protected] (G.R.) 2 Clinic of Obstetrics and Gynecology, San Gerardo Hospital, 20900 Monza, Italy 3 Department of Biochemistry and Biomedicine, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RH, UK; [email protected] 4 Division of Pathology, San Gerardo Hospital, 20900 Monza, Italy; [email protected] 5 Medical Genetics Laboratory, San Gerardo Hospital, 20900 Monza, Italy; [email protected] (E.G.); [email protected] (E.S.) Citation: Conconi, D.; Redaelli, S.; * Correspondence: [email protected] (D.C.); [email protected] (A.B.); Lissoni, A.A.; Cilibrasi, C.; Perego, P.; Tel.: +39-0264488133 (A.B.) Gautiero, E.; Sala, E.; Paderno, M.; † Co-first authorship. Dalprà, L.; Landoni, F.; et al. Genomic and Epigenomic Profile of Uterine Abstract: Uterine smooth muscle tumors of uncertain malignant potential (STUMPs) represent a Smooth Muscle Tumors of Uncertain heterogeneous group of tumors that cannot be histologically diagnosed as unequivocally benign Malignant Potential (STUMPs) or malignant. -

Molecular Alterations in Bone Development and Bone
MOLECULAR ALTERATIONS IN BONE DEVELOPMENT AND BONE TUMORIGENESIS DISSERTATION Presented in Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Graduate School of The Ohio State University By Emilia Mahoney, M.D. Molecular, Cellular and Developmental Biology Graduate Program The Ohio State University 2009 Dissertation Committee: Lawrence Kirschner, M.D., PhD, Advisor Rebecca Jackson, M.D. Michael Ostrowski, PhD Thomas Rosol, D.V.M., PhD Copyright by Emilia Mahoney 2009 ABSTRACT Bone is a specialized connective tissue that forms, together with the cartilage, the skeletal system. These tissues play important roles in locomotion, protection of vital organs and in metabolism. The fundamental constituents in bone are the extracellular matrix and the cells. The extracellular matrix is formed by collagen fibers and noncollagenous proteins. The bone cells are represented by osteoblasts, osteocytes and osteoclasts. There are many genetic alterations that can affect either the developing bone leading to skeletal deformities or the mature bone, leading to a vast array of pathological entities among which the tumors represent a significant percentage. The work presented here is focused on both these aspects by studying Gremlin regulation during limb development and Prkar1a mutation that causes formation of unique bone tumors through PKA dysregulation. Chapter 2 comprises a study on the developmental failure of distal limb structures seen in the mouse limb deformity ( ld ) phenotype. This anomaly is caused by the loss of Gremlin in the limb buds either through mutation of Grem1 or by loss of a transcriptional global control region (GCR) located in the neighboring Fmn1 gene. This study describes a new allele of ld due to complete deletion of Fmn1 , including its GCR, thus providing information about the role of this transcriptional global control region in Gremlin expression as well as long-range transcriptional effects that extend beyond Fmn1 and Grem1.