Abel Ecology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
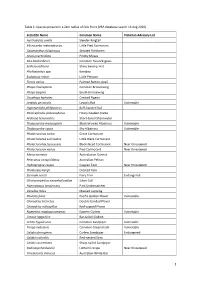
Scientific Name Common Name Victorian A
Table 1: Species present in a 2km radius of Crib Point (VBA database search 13 Aug 2020) Scientific Name Common Name Victorian Advisory List Austrolestes analis Slender Ringtail Microcarbo melanoleucos Little Pied Cormorant Calamanthus fuliginosus Striated Fieldwren Acacia verticillata Prickly Moses Poa labillardierei Common Tussock-grass Selliera radicans Shiny Swamp-mat Phyllostachys spp. Bamboo Eudyptula minor Little Penguin Turnix varius Painted Button-quail Phaps chalcoptera Common Bronzewing Phaps elegans Brush Bronzewing Ocyphaps lophotes Crested Pigeon Lewinia pectoralis Lewin's Rail Vulnerable Hypotaenidia philippensis Buff-banded Rail Poliocephalus poliocephalus Hoary-headed Grebe Ardenna tenuirostris Short-tailed Shearwater Thalassarche melanophris Black-browed Albatross Vulnerable Thalassarche cauta Shy Albatross Vulnerable Phalacrocorax carbo Great Cormorant Phalacrocorax sulcirostris Little Black Cormorant Phalacrocorax fuscescens Black-faced Cormorant Near threatened Phalacrocorax varius Pied Cormorant Near threatened Morus serrator Australasian Gannet Pelecanus conspicillatus Australian Pelican Hydroprogne caspia Caspian Tern Near threatened Thalasseus bergii Crested Tern Sternula nereis Fairy Tern Endangered Chroicocephalus novaehollandiae Silver Gull Haematopus longirostris Pied Oystercatcher Vanellus miles Masked Lapwing Pluvialis fulva Pacific Golden Plover Vulnerable Charadrius bicinctus Double-banded Plover Charadrius ruficapillus Red-capped Plover Numenius madagascariensis Eastern Curlew Vulnerable Limosa lapponica -

Networks in a Large-Scale Phylogenetic Analysis: Reconstructing Evolutionary History of Asparagales (Lilianae) Based on Four Plastid Genes
Networks in a Large-Scale Phylogenetic Analysis: Reconstructing Evolutionary History of Asparagales (Lilianae) Based on Four Plastid Genes Shichao Chen1., Dong-Kap Kim2., Mark W. Chase3, Joo-Hwan Kim4* 1 College of Life Science and Technology, Tongji University, Shanghai, China, 2 Division of Forest Resource Conservation, Korea National Arboretum, Pocheon, Gyeonggi- do, Korea, 3 Jodrell Laboratory, Royal Botanic Gardens, Kew, Richmond, United Kingdom, 4 Department of Life Science, Gachon University, Seongnam, Gyeonggi-do, Korea Abstract Phylogenetic analysis aims to produce a bifurcating tree, which disregards conflicting signals and displays only those that are present in a large proportion of the data. However, any character (or tree) conflict in a dataset allows the exploration of support for various evolutionary hypotheses. Although data-display network approaches exist, biologists cannot easily and routinely use them to compute rooted phylogenetic networks on real datasets containing hundreds of taxa. Here, we constructed an original neighbour-net for a large dataset of Asparagales to highlight the aspects of the resulting network that will be important for interpreting phylogeny. The analyses were largely conducted with new data collected for the same loci as in previous studies, but from different species accessions and greater sampling in many cases than in published analyses. The network tree summarised the majority data pattern in the characters of plastid sequences before tree building, which largely confirmed the currently recognised phylogenetic relationships. Most conflicting signals are at the base of each group along the Asparagales backbone, which helps us to establish the expectancy and advance our understanding of some difficult taxa relationships and their phylogeny. -

Cunninghamia Date of Publication: 18/11/2013 a Journal of Plant Ecology for Eastern Australia
Cunninghamia Date of Publication: 18/11/2013 A journal of plant ecology for eastern Australia ISSN 0727- 9620 (print) • ISSN 2200 - 405X (Online) Impact of broom, Cytisus scoparius (Fabaceae), in naturally treeless sub-alpine frost-hollow vegetation communities at the Barrington Tops, south-eastern Australia John R. Hosking1,3, Mellesa Schroder2 and Bruce McCorkell1 1Tamworth Agricultural Institute, NSW Department of Primary Industries, 4 Marsden Park Road, Calala, NSW 2340 AUSTRALIA. email [email protected] 2NSW Office of Environment and Heritage, National Parks and Wildlife Service, corner of Kosciusko Way and Thredbo Terrace, Jindabyne, NSW 2627 AUSTRALIA. email [email protected] 3current address: NCW Beadle Herbarium, University of New England, Armidale, NSW 2351 AUSTRALIA. email [email protected] Abstract: The exotic shrub Cytisus scoparius (L.) Link (family Fabaceae), known as broom, is having a major impact on native vegetation in naturally treeless sub-alpine frost-hollow areas (c. 32o 01’ 37” S, 151o 26’ 12” E’, 1440 m elevation) at the Barrington Tops, New South Wales, in south-eastern Australia. This vegetation is of limited extent and has significant biogeographical and ecological importance. Nine paired 10 m line transects were compared, with one of the pair in areas of almost 100% Cytisus scoparius and the other in adjacent areas without Cytisus scoparius. Results were compared with species recorded from this area in the 1930s. There are obvious differences in native vegetation in areas with and without Cytisus scoparius. On average there were 5.1 (range 2–10) species per 10 m in areas of almost 100% Cytisus scoparius cover and 17.0 (12–25) species per 10 m in areas adjacent to infested areas but without Cytisus scoparius. -

The Vegetation Communities Dry Eucalypt Forest and Woodland
Edition 2 From Forest to Fjaeldmark The Vegetation Communities Dry eucalypt forest and woodland Eucalyptus amygdalina Edition 2 From Forest to Fjaeldmark 1 Dry eucalypt forest and woodland Community (Code) Page Eucalyptus amygdalina coastal forest and woodland (DAC) 11 Eucalyptus amygdalina forest and woodland on dolerite (DAD) 13 Eucalyptus amygdalina forest and woodland on sandstone (DAS) 15 Eucalyptus amygdalina forest on mudstone (DAM) 17 Eucalyptus amygdalina inland forest and woodland on Cainozoic deposits (DAZ) 19 Eucalyptus amygdalina–Eucalyptus obliqua damp sclerophyll forest (DSC) 22 Eucalyptus barberi forest and woodland (DBA) 24 Eucalyptus coccifera forest and woodland (DCO) 25 Eucalyptus cordata forest (DCR) 27 Eucalyptus dalrympleana–Eucalyptus pauciflora forest and woodland (DDP) 29 Eucalyptus delegatensis dry forest and woodland (DDE) 31 Eucalyptus globulus dry forest and woodland (DGL) 33 Eucalyptus gunnii woodland (DGW) 35 Eucalyptus morrisbyi forest and woodland (DMO) 37 Eucalyptus nitida dry forest and woodland (DNI) 39 Eucalyptus nitida Furneaux forest (DNF) 41 Eucalyptus obliqua dry forest (DOB) 43 Eucalyptus ovata forest and woodland (DOV) 45 Eucalyptus ovata heathy woodland (DOW) 48 Eucalyptus pauciflora forest and woodland not on dolerite (DPO) 50 Eucalyptus pauciflora forest and woodland on dolerite (DPD) 52 Eucalyptus perriniana forest and woodland (DPE) 54 Eucalyptus pulchella forest and woodland (DPU) 56 Eucalyptus risdonii forest and woodland (DRI) 58 Eucalyptus rodwayi forest and woodland (DRO) 60 Eucalyptus -

Flora of Australia, Volume 46, Iridaceae to Dioscoreaceae
FLORA OF AUSTRALIA Volume 46 Iridaceae to Dioscoreaceae This volume was published before the Commonwealth Government moved to Creative Commons Licensing. © Commonwealth of Australia 1986. This work is copyright. You may download, display, print and reproduce this material in unaltered form only (retaining this notice) for your personal, non-commercial use or use within your organisation. Apart from any use as permitted under the Copyright Act 1968, no part may be reproduced or distributed by any process or stored in any retrieval system or data base without prior written permission from the copyright holder. Requests and inquiries concerning reproduction and rights should be addressed to: [email protected] FLORA OF AUSTRALIA The nine families in this volume of the Flora of Australia are Iridaceae, Aloeaceae, Agavaceae, Xanthorrhoeaceae, Hanguan- aceae, Taccaceae, Stemonaceae, Smilacaceae and Dioscoreaceae. The Xanthorrhoeaceae has the largest representation with 10 genera and 99 species. Most are endemic with a few species of Lomandra and Romnalda extending to neighbouring islands. The family includes the spectacular blackboys and grass-trees. The Iridaceae is largely represented by naturalised species with 52 of the 78 species being introduced. Many of the introductions are ornamentals and several have become serious weeds. Patersonia is the largest genus with all 17 species endemic. Some of these are cultivated as ornamentals. The Dioscoreaccae is a family of economic significance, particularly in the old world tropics where some species are cultivated or collected for their tubers and bulbils. In Australia there are 5 species, one of which is a recent introduction. The endemic and native species, commonly known as yams, are traditionally eaten by the Aborigines. -

Molecular Systematics of the Lomandra Labill. Complex
Molecular Systematics of the Lomandra Labill. Complex (Asparagales: Laxmanniaceae) by Matthew J. Donnon, B.Sc.Hons A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy Ecology and Evolutionary Biology Faculty of Science The University of Adelaide September 2009 1. General Introduction 1.1 General Introduction Laxmanniaceae (Monocotyledonae: Asparagales) are a tropical to temperate family of 14 genera with approximately 180 species from Australasia, SE Asia, the Mascarenes, New Caledonia, New Guinea, New Zealand, North and South America and the Pacific Islands (Conran 1998). Nevertheless, they are primarily an Australian family with many species spread across the continent, surviving and flourishing in an extraordinary variety of environments ranging from the monsoon tropics and subtropical rainforest to temperate Mediterranean heathlands and coastal sand dunes. Laxmanniaceae have a long historical record, with fossilised leaves of Paracordyline Conran species recognised from Eocene South Australia (Conran and Christophel, 1998), Oligocene Kerguélen Islands (Conran, 1997a) and Cordyline Comm. ex R.Br. pollen identified in Lower Miocene New Zealand sediments (Couper, 1953). Molecular dating work by Janssen and Bremer (2004) based on rbcL sequence data estimated the divergence origin of the Asparagaceae sensu lato (a wider family which includes Laxmanniaceae and thus Lomandra) at approximately 90 million years ago using nonparametric rate smoothing. This age estimation was given additional support by further work incorporating geological area cladogram comparisons and dispersal-vicariance analysis (Bremer and Janssen 2006). This latter study also indicated a South Gondwanan origin for the group. The plants are generally rhizomatous or tufted-caespitose, but the different species and genera are highly variable in size and morphology. -

Lomandra Hystrix (R.Br.) L.R.Fraser & Vickery Family: Asparagaceae Fraser, L.R
Australian Tropical Rainforest Plants - Online edition Lomandra hystrix (R.Br.) L.R.Fraser & Vickery Family: Asparagaceae Fraser, L.R. & Vickery, J.W. (1937) Proceedings of the Linnean Society of New South Wales 62: 286. Common name: Creek Mat-Rush; Mat-Rush, Creek; Matrush Stem Usually flowers and fruits as a shrubby plant about 1 m tall. Leaves Leaf blades linear, about 80-100 x 1-2 cm. Margin smooth except for 1 or 2 teeth below the apex. Bases sheathing, brown and scarious. Venation longitudinal and parallel. Flowers Inflorescence about as long as the leaves or shorter than the leaves. Male flowers: Flowers sessile, pleasantly perfumed, about 3 mm diam., arranged in whorls on the main branches of the inflorescence. Stamens six, attached to the tepals. Anthers yellow. Female flowers: Staminodes Flowers. © Barry Jago dimorphic, the short ones alternating with the inner tepals and the longer ones opposite the inner tepals. Ovary green, stigma 3-lobed. Fruit Infructescence with many long spiny bracts, each bract about 1-4 cm long. Fruits globular, about 5 x 4 mm, 3-lobed in transverse section. Style persistent at the apex of each fruit. Seeds up to three per fruit, each seed about 4 mm long. Testa very thin. Endosperm hard and starchy. Embryo slightly curved, about 1.8 mm long. Seedlings Seeds retained on threads about 4-8 mm long. Leaves radical, venation parallel and margin smooth. First leaf blade linear, about 17-40 x 1-1.5 mm, second leaf blade about 40-60 mm long. At the tenth Scale bar 10mm. -

And Type the TITLE of YOUR WORK in All Caps
PHYLOGENOMIC PLACEMENT OF ANCIENT POLYPLOIDY EVENTS WITHIN THE POALES AND AGAVOIDEAE (ASPARAGALES) by MICHAEL RAMON MCKAIN (Under the Direction of James H. Leebens-Mack) ABSTRACT Polyploidy has been an important component to the evolution of angiosperms. Recent studies have shown that an ancient polyploid (paleopolyploid) event can be traced to the lineage leading to the diversification of all angiosperms, and it has long been known that recurring polyploid events can be found throughout the angiosperm tree of life. With the advent of high- throughput sequencing, the prominent place of paleopolyploid events in the evolutionary history of angiosperms has become increasingly clear. Polyploidy is thought to spur both diversification and trait innovation through the duplication and reworking of gene networks. Understanding the evolutionary impact of paleopolyploidy within the angiosperms requires knowing when these events occurred during angiosperm evolution. This study utilizes a high-throughput phylogenomic approach to identify the timing of paleopolyploid events by comparing the origin of paralogous genes within a gene family to a known species tree. Transcriptome data derived from taxa in lineages with previously little to no genomic data, were utilized to assess the timing of duplication events within hundreds of gene families. Previously described paleopolyploid events in the history of grasses, identified through analyses of syntenic blocks within Poaceae genomes, were placed on the Poales phylogeny and the implications of these events were considered. Additionally, a previously unverified paleopolyploidy event was found to have occurred in a common ancestor of all members of the Asparagales and commelinids (including Poales, Zingiberales, Commelinales, Arecales and Dasypogonales). The phylogeny of the Asparagaceae subfamily Agavoideae was resolved using whole chloroplast genomes, and two previously unknown paleopolyploid events were described within the context of that phylogeny. -

Southern Forest Life Plant List
Southern Forest Life Botanical Inventory Regeneration after the Border Fire of January 5, 2020 FIRST MODE OF FAMILY GENUS SPECIES SUB-SPECIES COMMON NAME REGENERATION REGROWTH APIACEAE Hydrocotyle laxiflora Snking Pennywort FeB 8 sproung APIACEAE Platysace lanceolata ShruBBy Platysace FeB 27 seedlings APIACEAE Xanthosia pilosa Woolly Xanthosia APIACEAE Xanthosia tridentata Hill Xanthosia APOCYNACEAE Marsdenia rostrata Common Milk-vine FeB 5 sproung APOCYNACEAE Tylophora barbata Bearded Tylophora FeB 8 sproung ARALIACEAE Polyscias sambucifolia Bipinnate leaves Ferny Panax ASPARAGACEAE (suBf. Eustrephus lafolius WomBat Berry Lomandroideae) FeB 29 sproung ASPARAGACEAE (suBf. Lomandra confer>folia leptostachya Lomandroideae) ASPARAGACEAE (suBf. Lomandra filiformis filiformis Lomandroideae) FeB 15 sproung ASPARAGACEAE (suBf. Lomandra glauca Pale Mat-rush Lomandroideae) FeB 15 sproung ASPARAGACEAE (suBf. Lomandra longifolia longifolia Spinyhead Mat-rush Lomandroideae) Jan 28 sproung ASPARAGACEAE (suBf. Lomandra mul>flora mul>flora Many-flowered Mat-rush Lomandroideae) FeB 27 sproung ASPARAGACEAE (suBf. Thysanotus juncifolius Branching Fringe Lily Lomandroideae) ASPLENIACEAE Asplenium flabellifolium Necklace Fern ASTERACEAE Cassinia longifolia Shining Cassinia FeB 27 sproung ASTERACEAE Coronidium elatum elatum Tall Everlas7ng ASTERACEAE Coronidium scorpioides Bu[on Everlas7ng FeB 27 sproung ASTERACEAE Cotula australis Carrot Weed Jan 28 sproung ASTERACEAE *Facelis retusa Annual Trampweed FeB 27 ASTERACEAE *Gamochaeta sp. Cudweed FeB 8 sproung ASTERACEAE -

Lomandra Hystrix Click on Images to Enlarge
Species information Abo ut Reso urces Hom e A B C D E F G H I J K L M N O P Q R S T U V W X Y Z Lomandra hystrix Click on images to enlarge Family Asparagaceae Scientific Name Lomandra hystrix (R.Br.) L.R.Fraser & Vickery Fraser, L.R. & Vickery, J.W. (1937) Proceedings of the Linnean Society of New South Wales 62: 286. Flowers. Copyright Barry Jago Common name Creek Mat-Rush; Mat-Rush, Creek; Matrush Stem Usually flowers and fruits as a shrubby plant about 1 m tall. Leaves Leaf blades linear, about 80-100 x 1-2 cm. Margin smooth except for 1 or 2 teeth below the apex. Bases sheathing, brown and scarious. Venation longitudinal and parallel. Scale bar 10mm. Copyright CSIRO Flowers Inflorescence about as long as the leaves or shorter than the leaves. Male flowers: Flowers sessile, pleasantly perfumed, about 3 mm diam., arranged in whorls on the main branches of the inflorescence. Stamens six, attached to the tepals. Anthers yellow. Female flowers: Staminodes dimorphic, the short ones alternating with the inner tepals and the longer ones opposite the inner tepals. Ovary green, stigma 3-lobed. Fruit Infructescence with many long spiny bracts, each bract about 1-4 cm long. Fruits globular, about 5 x 4 mm, 3- lobed in transverse section. Style persistent at the apex of each fruit. Seeds up to three per fruit, each seed about 4 mm long. Testa very thin. Endosperm hard and starchy. Embryo slightly curved, about 1.8 mm long. -

5, and J. Chris Pires
American Journal of Botany 99(2): 330–348. 2012. Q UALITY AND QUANTITY OF DATA RECOVERED FROM MASSIVELY PARALLEL SEQUENCING: EXAMPLES IN 1 ASPARAGALES AND POACEAE P . R OXANNE S TEELE 2 , K ATE L. HERTWECK 3 , D USTIN M AYFIELD 4 , M ICHAEL R. MCKAIN 5 , J AMES L EEBENS-MACK 5 , AND J. CHRIS P IRES 3,6 2 Department of Biology, 6001 W. Dodge Street, University of Nebraska at Omaha, Omaha, Nebraska 68182-0040 USA; 3 National Evolutionary Synthesis Center, 2024 W. Main Street, Suite A200, Durham, North Carolina 27705-4667 USA; 4 Biological Sciences, 1201 Rollins St., Bond LSC 311, University of Missouri, Columbia, Missouri 65211 USA; and 5 Plant Biology, 4504 Miller Plant Sciences, University of Georgia, Athens, Georgia 30602 USA • Premise of the study: Genome survey sequences (GSS) from massively parallel sequencing have potential to provide large, cost-effective data sets for phylogenetic inference, replace single gene or spacer regions as DNA barcodes, and provide a plethora of data for other comparative molecular evolution studies. Here we report on the application of this method to estimat- ing the molecular phylogeny of core Asparagales, investigating plastid gene losses, assembling complete plastid genomes, and determining the type and quality of assembled genomic data attainable from Illumina 80 – 120-bp reads. • Methods: We sequenced total genomic DNA from samples in two lineages of monocotyledonous plants, Poaceae and Aspara- gales, on the Illumina platform in a multiplex arrangement. We compared reference-based assemblies to de novo contigs, evaluated consistency of assemblies resulting from use of various references sequences, and assessed our methods to obtain sequence assemblies in nonmodel taxa. -

Hutchinson (Agavales) Vs. Huber Y Dahlgren (Asparagales)
Boletín de la Sociedad Botánica de México 56: 45-56, 1995 DOI: 10.17129/botsci.1463 Bol. Soc. Bot. México 56: 45-56 (1995) Hutchinson (Agavales) vs. Huber y Dahlgren (Asparagales): análisis moleculares sobre la filogenia y evolución de la familia Agavaceae sensu Hutchinson dentro de las monocotiledóneas LUIS E. EGUIARTE Departamento de Ecología Evolutiva, Centro de Ecología, Universidad Nacional Autónoma de México, Apartado Postal 70-275, C.P. 04510, D.F., México. Resumen: Para explorar diferentes ideas sobre la filogenia de las monocotiledóneas, y en particular sobre las Agavaceae se ns u Hutchinson (1934, 1959) y su relación con grupos emparentados dentro de las Asparagales, se analizó la secuencia de ADN del gen del cloroplasto rbcL para 134 especies (118 monocotiledóneas, empleando 16 dicotiledóneas paleohierbas como grupo externo). Se llevaron a cabo análisis de parsimonia utilizando métodos de distancia (neighbor-joining y UPGMA) y de máxima verosimilitud. Las filogenias muestran que Acorus calamus es la representante actual más cercana a las monocotiledóneas ancestrales. Los diferentes análisis identifican tres principales linajes evolutivos en las Agavaceae sensu Hutchinson: 1) El de las Agavaceae sensu stricto (Agave, Man/reda, Beschon ~ eria, Hesperaloe y Yucca), Hasta (Funkiaceae) y Chlorophytum (Anthericaceae). 2) El grupo que incluye a las Nolinaceae (Nolina, Beaucarnea y Dasylirion), a Sansevieria (Dracaenaceae) y a Danae (Rúscaceae). 3) El formado por las Asphodelaceae (Aloe, Haworthia y Kniphofia) y Dianella (Phormiaceae). Estos linajes contienen a la mayoría de las Asparagales tal como fueron definidas por Huber (1969) y Dahlgren et al. (1985). También se estimó un reloj molecular para obtener las fechas de divergencia entre los grupos relacionados con las Agavaceae.