Time-Course Microarray Analysis for Identifying Candidate Genes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differential Proteomic Analysis of the Pancreas of Diabetic Db/Db Mice Reveals the Proteins Involved in the Development of Complications of Diabetes Mellitus
Int. J. Mol. Sci. 2014, 15, 9579-9593; doi:10.3390/ijms15069579 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Differential Proteomic Analysis of the Pancreas of Diabetic db/db Mice Reveals the Proteins Involved in the Development of Complications of Diabetes Mellitus Victoriano Pérez-Vázquez 1,*, Juan M. Guzmán-Flores 1, Daniela Mares-Álvarez 1, Magdalena Hernández-Ortiz 2, Maciste H. Macías-Cervantes 1, Joel Ramírez-Emiliano 1 and Sergio Encarnación-Guevara 2 1 Depto. de Ciencias Médicas, División de Ciencias de la Salud, Campus León, Universidad de Guanajuato, León, Guanajuato 37320, Mexico; E-Mails: [email protected] (J.M.G.-F.); [email protected] (D.M.-A.); [email protected] (M.H.M.-C.); [email protected] (J.R.-E.) 2 Centro de Ciencias Genómicas, Universidad Nacional Autónoma de México, Cuernavaca, Morelos 62210, Mexico; E-Mails: [email protected] (M.H.-O.); [email protected] (S.E.-G.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +52-477-7143-812; Fax: +52-477-7167-623. Received: 4 April 2014; in revised form: 14 May 2014 / Accepted: 19 May 2014 / Published: 30 May 2014 Abstract: Type 2 diabetes mellitus is characterized by hyperglycemia and insulin-resistance. Diabetes results from pancreatic inability to secrete the insulin needed to overcome this resistance. We analyzed the protein profile from the pancreas of ten-week old diabetic db/db and wild type mice through proteomics. Pancreatic proteins were separated in two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and significant changes in db/db mice respect to wild type mice were observed in 27 proteins. -

ONLINE SUPPLEMENTARY TABLE Table 2. Differentially Expressed
ONLINE SUPPLEMENTARY TABLE Table 2. Differentially Expressed Probe Sets in Livers of GK Rats. A. Immune/Inflammatory (67 probe sets, 63 genes) Age Strain Probe ID Gene Name Symbol Accession Gene Function 5 WKY 1398390_at small inducible cytokine B13 precursor Cxcl13 AA892854 chemokine activity; lymph node development 5 WKY 1389581_at interleukin 33 Il33 BF390510 cytokine activity 5 WKY *1373970_at interleukin 33 Il33 AI716248 cytokine activity 5 WKY 1369171_at macrophage stimulating 1 (hepatocyte growth factor-like) Mst1; E2F2 NM_024352 serine-throenine kinase; tumor suppression 5 WKY 1388071_x_at major histocompatability antigen Mhc M24024 antigen processing and presentation 5 WKY 1385465_at sialic acid binding Ig-like lectin 5 Siglec5 BG379188 sialic acid-recognizing receptor 5 WKY 1393108_at major histocompatability antigen Mhc BM387813 antigen processing and presentation 5 WKY 1388202_at major histocompatability antigen Mhc BI395698 antigen processing and presentation 5 WKY 1371171_at major histocompatability antigen Mhc M10094 antigen processing and presentation 5 WKY 1370382_at major histocompatability antigen Mhc BI279526 antigen processing and presentation 5 WKY 1371033_at major histocompatability antigen Mhc AI715202 antigen processing and presentation 5 WKY 1383991_at leucine rich repeat containing 8 family, member E Lrrc8e BE096426 proliferation and activation of lymphocytes and monocytes. 5 WKY 1383046_at complement component factor H Cfh; Fh AA957258 regulation of complement cascade 4 WKY 1369522_a_at CD244 natural killer -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -

( 12 ) United States Patent
US010245229B2 (12 ) United States Patent ( 10 ) Patent No. : US 10 , 245 ,229 B2 Heartlein et al. (45 ) Date of Patent: Apr. 2 , 2019 ( 54 ) PULMONARY DELIVERY OF MRNA TO ( 56 ) References Cited NON -LUNG TARGET CELLS U . S . PATENT DOCUMENTS ( 71 ) Applicants : Shire Human Genetic Therapies, Inc. , 5 ,705 , 385 A 1 / 1998 Bally et al . Lexington , MA (US ) ; Ethris GmbH , 5 , 965, 434 A 10 / 1999 Wolff et al . Martinsried (DE ) 5 , 976 , 567 A 11 / 1999 Wheeler et al. 5 , 981 ,501 A 11/ 1999 Wheeler et al. 6 ,077 ,835 A 6 / 2000 Hanson et al . (72 ) Inventors : Michael Heartlein , Lexington , MA 6 ,534 , 484 B1 3 / 2003 Wheeler et al . (US ) ; Braydon Charles Guild , 6 ,734 , 171 B1 5 /2004 Saravolac et al. Lexington , MA (US ) ; Frank DeRosa , 6 , 815 ,432 B2 11/ 2004 Wheeler et al . Lexington , MA (US ) ; Carsten 7 , 422 , 902 B1 9 / 2008 Wheeler et al . Rudolph , Krailling ( DE ) ; Christian 7 ,745 ,651 B2 6 /2010 Heyes et al. 7 ,799 , 565 B2 9 /2010 MacLachlan et al. Plank , Wessling (DE ) 7 , 803 ,397 B2 9 / 2010 Heyes et al . 7 , 901, 708 B2 3 / 2011 MacLachlan et al. (73 ) Assignees : Translate Bio , Inc . , Lexington , MA 8 , 101 ,741 B2 1 / 2012 MacLachlan et al . (US ) ; Ethris GmbH , Martinsried (DE ) 8 , 188 , 263 B2 5 / 2012 MacLachlan et al . 8 , 236 , 943 B2 8 / 2012 Lee et al. 8 ,329 ,070 B2 12 /2012 MacLachlan et al . ( * ) Notice : Subject to any disclaimer , the term of this 8 ,513 ,403 B2 8 / 2013 MacLachlan et al. -

A Genomic Analysis of Rat Proteases and Protease Inhibitors
A genomic analysis of rat proteases and protease inhibitors Xose S. Puente and Carlos López-Otín Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, 33006-Oviedo, Spain Send correspondence to: Carlos López-Otín Departamento de Bioquímica y Biología Molecular Facultad de Medicina, Universidad de Oviedo 33006 Oviedo-SPAIN Tel. 34-985-104201; Fax: 34-985-103564 E-mail: [email protected] Proteases perform fundamental roles in multiple biological processes and are associated with a growing number of pathological conditions that involve abnormal or deficient functions of these enzymes. The availability of the rat genome sequence has opened the possibility to perform a global analysis of the complete protease repertoire or degradome of this model organism. The rat degradome consists of at least 626 proteases and homologs, which are distributed into five catalytic classes: 24 aspartic, 160 cysteine, 192 metallo, 221 serine, and 29 threonine proteases. Overall, this distribution is similar to that of the mouse degradome, but significatively more complex than that corresponding to the human degradome composed of 561 proteases and homologs. This increased complexity of the rat protease complement mainly derives from the expansion of several gene families including placental cathepsins, testases, kallikreins and hematopoietic serine proteases, involved in reproductive or immunological functions. These protease families have also evolved differently in the rat and mouse genomes and may contribute to explain some functional differences between these two closely related species. Likewise, genomic analysis of rat protease inhibitors has shown some differences with the mouse protease inhibitor complement and the marked expansion of families of cysteine and serine protease inhibitors in rat and mouse with respect to human. -

Uniprot Nr. Proseek Panel 2,4-Dienoyl-Coa Reductase, Mitochondrial
Protein Name (Short Name) Uniprot Nr. Proseek Panel 2,4-dienoyl-CoA reductase, mitochondrial (DECR1) Q16698 CVD II 5'-nucleotidase (5'-NT) P21589 ONC II A disintegrin and metalloproteinase with thrombospondin motifs 13 (ADAM-TS13) Q76LX8 CVD II A disintegrin and metalloproteinase with thrombospondin motifs 15 (ADAM-TS 15) Q8TE58 ONC II Adenosine Deaminase (ADA) P00813 INF I ADM (ADM) P35318 CVD II ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 (CD38) P28907 NEU I Agouti-related protein (AGRP) O00253 CVD II Alpha-2-macroglobulin receptor-associated protein (Alpha-2-MRAP) P30533 NEU I Alpha-L-iduronidase (IDUA) P35475 CVD II Alpha-taxilin (TXLNA) P40222 ONC II Aminopeptidase N (AP-N) P15144 CVD III Amphiregulin (AR) P15514 ONC II Angiopoietin-1 (ANG-1) Q15389 CVD II Angiopoietin-1 receptor (TIE2) Q02763 CVD II Angiotensin-converting enzyme 2 (ACE2) Q9BYF1 CVD II Annexin A1 (ANXA1) P04083 ONC II Artemin (ARTN) Q5T4W7 INF I Axin-1 (AXIN1) O15169 INF I Azurocidin (AZU1 P20160 CVD III BDNF/NT-3 growth factors receptor (NTRK2) Q16620 NEU I Beta-nerve growth factor (Beta-NGF) P01138 NEU I, INF I Bleomycin hydrolase (BLM hydrolase) Q13867 CVD III Bone morphogenetic protein 4 (BMP-4) P12644 NEU I Bone morphogenetic protein 6 (BMP-6) P22004 CVD II Brain-derived neurotrophic factor (BDNF) P23560 NEU I, INF I Brevican core protein (BCAN) Q96GW7 NEU I Brorin (VWC2) Q2TAL6 NEU I Brother of CDO (Protein BOC) Q9BWV1 CVD II Cadherin-3 (CDH3) P22223 NEU I Cadherin-5 (CDH5) P33151 CVD III Cadherin-6 (CDH6) P55285 NEU I Carbonic anhydrase 5A, mitochondrial -

Mrna Expression in Human Leiomyoma and Eker Rats As Measured by Microarray Analysis
Table 3S: mRNA Expression in Human Leiomyoma and Eker Rats as Measured by Microarray Analysis Human_avg Rat_avg_ PENG_ Entrez. Human_ log2_ log2_ RAPAMYCIN Gene.Symbol Gene.ID Gene Description avg_tstat Human_FDR foldChange Rat_avg_tstat Rat_FDR foldChange _DN A1BG 1 alpha-1-B glycoprotein 4.982 9.52E-05 0.68 -0.8346 0.4639 -0.38 A1CF 29974 APOBEC1 complementation factor -0.08024 0.9541 -0.02 0.9141 0.421 0.10 A2BP1 54715 ataxin 2-binding protein 1 2.811 0.01093 0.65 0.07114 0.954 -0.01 A2LD1 87769 AIG2-like domain 1 -0.3033 0.8056 -0.09 -3.365 0.005704 -0.42 A2M 2 alpha-2-macroglobulin -0.8113 0.4691 -0.03 6.02 0 1.75 A4GALT 53947 alpha 1,4-galactosyltransferase 0.4383 0.7128 0.11 6.304 0 2.30 AACS 65985 acetoacetyl-CoA synthetase 0.3595 0.7664 0.03 3.534 0.00388 0.38 AADAC 13 arylacetamide deacetylase (esterase) 0.569 0.6216 0.16 0.005588 0.9968 0.00 AADAT 51166 aminoadipate aminotransferase -0.9577 0.3876 -0.11 0.8123 0.4752 0.24 AAK1 22848 AP2 associated kinase 1 -1.261 0.2505 -0.25 0.8232 0.4689 0.12 AAMP 14 angio-associated, migratory cell protein 0.873 0.4351 0.07 1.656 0.1476 0.06 AANAT 15 arylalkylamine N-acetyltransferase -0.3998 0.7394 -0.08 0.8486 0.456 0.18 AARS 16 alanyl-tRNA synthetase 5.517 0 0.34 8.616 0 0.69 AARS2 57505 alanyl-tRNA synthetase 2, mitochondrial (putative) 1.701 0.1158 0.35 0.5011 0.6622 0.07 AARSD1 80755 alanyl-tRNA synthetase domain containing 1 4.403 9.52E-05 0.52 1.279 0.2609 0.13 AASDH 132949 aminoadipate-semialdehyde dehydrogenase -0.8921 0.4247 -0.12 -2.564 0.02993 -0.32 AASDHPPT 60496 aminoadipate-semialdehyde -
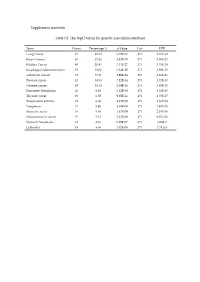
Supplement Materials Table S1. the Top15 Terms for Genetic Association
Supplement materials Table S1. The top15 terms for genetic association database. Term Count Percentage % p-Value List FDR Lung Cancer 73 25.34 6.70E-36 271 1.16E-32 Breast Cancer 67 23.26 2.92E-30 271 5.06E-27 Bladder Cancer 60 20.83 3.33E-27 271 5.78E-24 Esophageal adenocarcinoma 52 18.06 6.24E-29 271 1.08E-25 Colorectal Cancer 51 17.71 4.99E-24 271 8.66E-21 Prostate cancer 42 14.58 7.12E-14 271 1.23E-10 Ovarian cancer 39 13.54 2.04E-16 271 3.89E-13 Pancreatic Neoplasms 20 6.94 1.45E-08 271 2.52E-05 Thyroid cancer 19 6.59 9.98E-11 271 1.73E-07 Rheumatoid arthritis 19 6.60 8.82E-08 271 1.53E-04 Neoplasms 17 5.90 4.58E-08 271 7.94E-05 Stomach cancer 16 5.56 1.17E-09 271 2.03E-06 Head and neck cancer 15 5.21 5.23E-09 271 9.07E-06 Stomach Neoplasms 14 4.86 6.20E-07 271 1.08E-3 Leukemia 14 4.86 1.02E-06 271 1.78 E-3 Table S2. The results of the reverse docking for AutoDock Vina. PDB Protein Gene_Name Uniprot_ID Score 4TVJ Poly [ADP-ribose] polymerase 2 PARP2 Q9UGN5 -10.80 2DQ7 Proto-oncogene tyrosine-protein kinase Fyn FYN P06241 -10.00 4KIK Inhibitor of nuclear factor kappa-B kinase IKBKB O14920 -10.00 subunit beta 3EQR Activated CDC42 kinase 1 ACK1/TNK2 Q07912 -9.90 3MTF Activin receptor type-1 ACVR1 Q04771 -9.90 4GV0 Poly [ADP-ribose] polymerase 3 PARP3 Q9Y6F1 -9.90 1M6I Programmed cell death protein 8 AIF O95831 -9.80 1UA2 Cell division protein kinase 7 CDK7 P50613 -9.80 1Z6T Apoptotic protease activating factor 1 APAF1 O14727 -9.80 1ZXM DNA topoisomerase II TOPII P11388 -9.80 2WOU Serine/Threonine-protein kinase NEK7 NEK7 Q8TDX7 -9.60 -

Structural Basis of the Resistance of an Insect Carboxypeptidase to Plant Protease Inhibitors
Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors Alex Baye´ s*, Mireia Comellas-Bigler†, Monica Rodrı´guez de la Vega*, Klaus Maskos†, Wolfram Bode†, Francesc X. Aviles*, Maarten A. Jongsma‡, Jules Beekwilder‡§, and Josep Vendrell*§ *Departament de Bioquı´mica i Biologia Molecular, Facultat de Cie` ncies, and Institut de Biotecnologia i de Biomedicina, Universitat Auto` noma de Barcelona, E-08193 Bellaterra, Spain; †Proteinase Research Group, Max Planck Institute of Biochemistry, D-82152 Martinsried, Germany; and ‡Plant Research International, Postbus 16, 6700, Wageningen, The Netherlands Edited by Clarence A. Ryan, Jr., Washington State University, Pullman, WA, and approved September 26, 2005 (received for review June 29, 2005) Corn earworm (Helicoverpa zea), also called tomato fruitworm, is zymes depending on the specific inhibitor repertoire encoun- a common pest of many Solanaceous plants. This insect is known tered in the host plant which they colonize (5, 6). Analyses of to adapt to the ingestion of plant serine protease inhibitors by trypsin-like enzymes have shown that PI-sensitive and PI- using digestive proteases that are insensitive to inhibition. We adapted trypsins differ only marginally in amino acid sequence have now identified a B-type carboxypeptidase of H. zea (CPBHz) and substrate specificity but exhibit huge differences in sensi- insensitive to potato carboxypeptidase inhibitor (PCI) in corn ear- tivity against PIs (7). However, in the absence of crystal struc- worm. To elucidate the structural features leading to the adapta- tures, there has been no conclusive mechanistic, structure-based tion of the insect enzyme, the crystal structure of the recombinant explanation for the apparent insensitivity of PI-adapted insect CPBHz protein was determined by x-ray diffraction. -

Mckinney, Clare Adele (2014) the Role of Angiotensin-(1-7) and Angiotensin-(1-9) in Vascular Remodelling
McKinney, Clare Adele (2014) The role of Angiotensin-(1-7) and Angiotensin-(1-9) in vascular remodelling. PhD thesis. http://theses.gla.ac.uk/5752/ Copyright and moral rights for this work are retained by the author A copy can be downloaded for personal non-commercial research or study, without prior permission or charge This work cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given Enlighten:Theses http://theses.gla.ac.uk/ [email protected] The Role of Angiotensin-(1-7) and Angiotensin-(1-9) in Vascular Remodelling Clare Adele McKinney BSc (Hons) Submitted in fulfilment of the requirements of the degree of Doctor of Philosophy in the Institute of Cardiovascular and Medical Sciences, University of Glasgow Institute of Cardiovascular and Medical Sciences College of Medical, Veterinary and Life Sciences University of Glasgow November 2014 © C.A McKinney 2014 ii Authors Declaration I declare that this thesis has been written entirely by myself and is a record of work performed by me with the exception of analysis of microRNA and target expression, and microRNA inhibitor studies which were performed in collaboration with Miss Lisa McArthur. The plasmid used to express the angiotensin type 2 receptor in cell lines was generated by Mr Daniel Villela, and experiments using this plasmid were performed in collaboration with Mr Daniel Villela. -

(12) United States Patent (10) Patent No.: US 8,993,295 B2 Seed Et Al
US008993295 B2 (12) United States Patent (10) Patent No.: US 8,993,295 B2 Seed et al. (45) Date of Patent: *Mar. 31, 2015 (54) METHODS, COMPOSITIONS, AND KITS FOR (56) References Cited THE SELECTIVE ACTIVATION OF PROTOXINS THROUGH COMBINATORIAL U.S. PATENT DOCUMENTS TARGETING 4,975,278 A 12/1990 Senter 5,156,840 A 10, 1992 Goers (75) Inventors: Brian Seed, Boston, MA (US); Jia Liu 6,258,360 B1 7/2001 Von Borstel 2002/0142359 A1 10/2002 Copley Wolfe, Winchester, MA (US); Glen S. 2003, OO54000 A1 3/2003 Dowdy Cho, Brookline, MA (US); Chia-Iun 2004/0048784 A1 3/2004 Keener et al. Tsai, Winchester, MA (US) 2009/00 16988 A1* 1/2009 Buckley ....................... 424/85.2 2010/0256070 A1* 10/2010 Seed et al. ................... 514, 19.3 (73) Assignee: The General Hospital Corporation, Boston, MA (US) FOREIGN PATENT DOCUMENTS WO WO98, 20135 A2 5, 1998 (*) Notice: Subject to any disclaimer, the term of this WO WOO1/14570 A1 3, 2001 patent is extended or adjusted under 35 WO WO 2004/094478 A2 11/2004 U.S.C. 154(b) by 1188 days. This patent is Subject to a terminal dis OTHER PUBLICATIONS claimer. Chiron et al. (JBC 272(50):31707-31711 (1997)).* Nygren et al., “Overview of the clinical efficacy of investigational (21) Appl. No.: 12/374,616 anticancer drugs” Journal of Internal Medicine. 253:46-75 (2003). Stenter et al., “Activation of prodrugs by antibody-enzyme conju (22) PCT Fled: Jul. 20, 2007 gates: a new approach to cancer therapy.” The FASEBJournal 4:188 193 (1990).