CICR Newsletter Template
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Fourth Report of Senior Pay and Perks in UK Universities History This
Transparency at the top? The fourth report of senior pay and perks in UK universities History This is the fourth report on pay and perks at the top of British higher education institutions (HEIs) to be published by the University and College Union (UCU). It forms part of the union’s ongoing campaign for greater transparency in higher education, including the rationale behind senior pay rises. UCU submitted a Freedom of Information (FoI) request to 158 HEIs in October 2017. This followed similar requests submitted in 2016, 2015 and 2014. All requests were designed to shine a light on the arbitrary nature of senior pay and perks in universities, and support the union’s call for reform. The basis for this report The FoI request that forms the basis of this report was sent to 158 (HEIs). It requested details of vice-chancellors’ (or head of institution if known by a different title) salaries and those of other senior post-holders earning over £100,000 at the institution during the academic year of 2016/17 (1 August 2016 to 31 July 2017). It also asked for details of flights, spending on hotels, spending on expenses and if the vice-chancellor was provided with accommodation by the university. Finally, we requested to know whether or not the vice-chancellor was a member of the remuneration committee, and requested a copy of the most recently ratified minutes of the institution’s remuneration committee. Variety of responses The questions on expenditure on flights, hotels, expenses and accommodation for vice-chancellors elicited a huge variation in responses with many institutions deploying exemptions under the Freedom of Information Act to avoid providing data. -

Enthusing Children About Chemistry Climate Change and Biogenic Emissions Predicting Properties Using Informatics the Oil Industr
Summer 2008 Enthusing children about chemistry Predicting properties using informatics Climate change and biogenic emissions The oil industry’s chemical challenges As I see it... So are you finding it increasingly diffi - Oil exploration doesn’t just offer a career for engineers – cult to attract good chemists? chemists are vital, too. Sarah Houlton spoke to Schlumberger’s It can be a challenge, yes. Many of the com - pany’s chemists are recruited here, and they Tim Jones about the crucial role of chemistry in the industry often move on to other sites such as Houston or Paris, but finding them in the first place can be a challenge. Maybe one reason is that the oil People don’t think of Schlumberger as We can’t rely on being able to find suitable industry doesn’t have the greatest profile in a chemistry-using company, but an chemistries in other industries, either, mainly chemistry, and people think it employs engi - engineering one. How important is because of the high temperatures and pressures neers, not chemists. But it’s something the chemistry in oil exploration? that we have to be able to work at. Typically, the upstream oil industry cannot manage without, It’s essential! There are many challenges for upper temperature norm is now 175°C, but even if they don’t realise it! For me, maintaining chemistry in helping to maintain or increase oil we’re increasingly looking to go over 200°C. – if not enhancing – our recruitment is perhaps production. It’s going to become increasingly For heavy oil, where we heat the oil up with one of the biggest issues we face. -

Womencount: Leaders in Higher Education 2016
WomenCount Leaders in Higher Education 2016 A report by Norma Jarboe OBE ‘There’s no magic about getting a good gender balance – just dogged repetition of what a high priority it is and a determination to seek out strong women candidates who could hold their own against any competition.’ WomenCount ‘If universities inhibit the progression of talented female staff, they in turn are unable to reach their full potential. We know that universities make a huge contribution to society through research, teaching and partnerships with businesses, among many other activities.’ Professor Dame Athene Donald, Professor of Experimental Physics and Master of Churchill College, University of Cambridge WomenCount are very grateful to Perrett Laver for their support, the Government Equalities Office for their enagagement and Imperial College London for hosting the launch of this report on 2 March 2016. Cover quotation: Sir Nicholas Montagu, Chair of Council, Queen Mary University of London. Published by WomenCount © March 2016, all rights reserved. www.women-count.org Designed and produced by Graffeg. WomenCount: Leaders in Higher Education 2016 Contents 3 Foreword 4 Executive summary 5 Introduction 6 Collective action 9 Collegial governance: diversity challenge or opportunity? 10 Governing bodies: women’s representation on the rise 11 Governing bodies: the balancing act 12 Chairs: few seats for women 14 Vice-Chancellors: barely a fifth are women 15 Chair and Vice-Chancellor teams 16 Executive teams: a pipeline of women leaders 17 Academic heads: less than a third are women 19 HEI income impact women’s leadership 21 Mapping Women’s Leadership in HEIs 22 Reflections on the research 27 The Index 37 Biographies of new Chairs 42 Biographies of new Vice-Chancellors 47 About WomenCount and the author 1 Foreword ‘Gender equality is not a matter of being nice to women. -

Perspective Lecture, Frankfurt February 14Th 2013 Professor Paul
Professor Paul Workman Bio: Perspective Lecture, Frankfurt February 14 th 2013 Professor Paul Workman – Short Biography Professor Paul Workman is a leader in the discovery and development of molecularly targeted cancer drugs and is a passionate advocate of personalized cancer therapy. A molecular pharmacologist and chemical biologist, Paul has been responsible for the discovery and development of a large number of new molecularly targeted cancer drugs. Paul is currently Deputy Chief Executive of The Institute of Cancer Research (ICR) and Director of the ICR’s Cancer Research UK Cancer Therapeutics Unit in Sutton, UK, which is the largest non-profit cancer drug discovery group worldwide. He is also Head of ICR’s Division of Cancer Therapeutics and Harrap Professor of Pharmacology and Therapeutics. Paul obtained his BSc (Hons) in Biochemistry from Leicester University (1973) and his PhD in Cancer Pharmacology from Leeds University (1977). He then moved to Cambridge to become a postdoctoral fellow and scientific staff member of the MRC Clinical Oncology Unit, MRC Centre, Cambridge University (1976-1990) where he established and led the cancer pharmacology group, making major contributions to drugs targeting tumour hypoxia. Following a period as UICC Visiting Fellow at Stanford University and SRI International, California (1989), Paul was appointed as Cancer Research Campaign Professor and Director of Laboratory Research in the Department of Medical Oncology, Beatson Laboratories, Glasgow University (1990-1993). Paul then gained experience in the pharmaceutical industry. From 1993-1997 Paul was Head of the Cancer Research Bioscience Section at AstraZeneca Pharmaceuticals, Alderley Park, UK where he also initiated and led the strategic alliance with Sugen. -

Situación Actual Y Perspectivas De Futuro Personalized Precision Oncology: Current Status and Future Perspectives Madrid, 21 Y 22 De Marzo / March 21-22, 2019
Simposio Internacional International Symposium Oncología personalizada de precisión: situación actual y perspectivas de futuro Personalized Precision Oncology: current status and future perspectives Madrid, 21 y 22 de marzo / March 21-22, 2019 BIO Paul Workman Professor Paul Workman FMedSci, FRS is Chief Executive and President of The Institute of Cancer Research (ICR). Professor Workman is a passionate advocate of personalised molecular medicine and is an enthusiastic practitioner of multidisciplinary cancer drug discovery and development approaches to 'drugging the cancer genome'. He also conceptualised the 'Pharmacological Audit Trail' approach. As well as establishing and successfully leading drug discovery project teams yielding clinical candidates, Professor Workman’s personal and collaborative research utilises molecular pharmacology and chemical biology approaches, including high-throughput and genome-wide as well as hypothesis- driven strategies, to interrogate cancer biology, identify and validate new drug targets, discover and develop chemical tools and drugs acting on these targets, identify predictive and mechanism of action biomarkers, and elucidate mechanisms of drug sensitivity of resistance. He is especially interested in exploiting the addictions, vulnerabilities and dependencies of cancer cells using a combination of small molecule tools and drugs alongside molecular genetic techniques. Professor Workman has successfully built a series of multidisciplinary drug discovery and development teams in the academic, large pharma and biotechnology company sectors. Through this experience he has been able to combine the best elements of each of these environments. He has been responsible for the discovery of a number of drug development candidates, including in particular pathfinding inhibitors of the HSP90 molecular chaperone and the PI3 kinase family of signalling enzymes. -
![Ecosystems for Innovation [Compatibility Mode]](https://docslib.b-cdn.net/cover/4930/ecosystems-for-innovation-compatibility-mode-1254930.webp)
Ecosystems for Innovation [Compatibility Mode]
Ecosystems for innovation Claire Ruskin CEO, Cambridge Network Innovation in Cambridge – how did it happen, how can we grow it and is it repeatable elsewhere? Material from Shai Vyakarnum – Judge Business School Tim Minshall – Institute for Manufacturing Claire Ruskin – CEO Cambridge Network Steven Ireland – East of England Inward Investment Cambridge Network – II EE www.cambridgenetwork.co.uk What’s special about Cambridge? The starting point … • A global ‘top five’ university: The University of Cambridge has 800 years at the top • Proximity to London and Europe : 5 international airports within 2 hours • Highly qualified employees: > 40% of people living in Cambridge having a high level qualification (compared to the national average of < 20%) • A few hi-tech companies back in the fifties • The start of a world class contract R&D cluster (the consultancies) from 1960 • => Evolving to a hi-tech cluster supporting > 143,000 jobs in the region. The cluster generates the equivalent of an NPV of £53bn in GDP. • => Good quality of life: Polls highlight Cambridgeshire as one of the best places to live in the UK • => Attitude: a good feeling about success and starting something new Cambridge Network – II EE www.cambridgenetwork.co.uk Why will Cambridge continue to have competitive advantage? • Diverse science base and research infrastructure, bringing excellent people to the Universities, business and medical organisations • Practice at innovation on demand as well as commercialisation • Collective learning and networking systems • Entrepreneurial -
BSCB Newsletter 2019A:BSCB Aut2k7
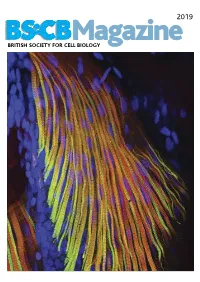
2019 BSCB Magazine BRITISH SOCIETY FOR CELL BIOLOGY 2019 CONTENTS BSCB Magazine News 2 Book reviews 8 Features 9 Meeting Reports 21 Summer students 25 Society Business 32 Editorial Front cover: microscopic Welcome to the 2019 BSCB Magazine! This year Mustafa Aydogan (University of Oxford), as well as to structure of pectoral fin and Susana and Stephen are filling in for our Newsletter BSCB postdoc poster of the year winners Dr Anna hypaxial muscles of a zebrafish Editor Ann Wheeler. We hope you will enjoy this Caballe (University of Oxford) and Dr Agata Gluszek- Danio rerio larvae at four days year’s magazine! Kustusz (University of Edinburgh). post fertilization. The immunostaining highlights the This year we had a number of fantastic one day In 2019, we will have our jointly BSCB-BSDB organization of fast (red) and meetings sponsored by BSCB. These focus meetings Spring meeting at Warwick University from 7th–10th slow (green) myosins. All nuclei are great way to meet and discuss your science with April, organised by BSCB members Susana Godinho are highlighted in blue (hoechst). experts in your field and to strengthen your network of and Vicky Sanz-Moreno. The programme for this collaborators within the UK. You can read more about meeting, which usually provides a broad spectrum of these meetings in the magazine. If you have an idea themes, has a focus on cancer biology: cell for a focus one day meeting, check how to apply for migration/invasion, organelle biogenesis, trafficking, funding on page 4. Our ambassadors have also been cell-cell communication. -

Alessio Ciulli
2015 Rising Stars Alessio Ciulli Alessio graduated in Chemistry (2002) from his hometown Florence under the late Ivano Bertini and obtained his PhD from the University of Cambridge (Chemistry, 2006), studying as a Gates Cambridge Scholar under the supervision of Chris Abell and in collaboration with Astex Pharmaceuticals. Following post-doctoral research on fragment-based drug design with Chris Abell and Tom Blundell, and an HFSP visiting Fellowship at Yale University to begin collaboration with Craig Crews (2009), he was awarded a BBSRC David Phillips Fellowship and returned to Cambridge to start his independent career in 2010. In 2013 Alessio was awarded an ERC Starting Grant and moved his laboratory to the School of Life Sciences at Dundee where he is currently an Associate Professor in Chemical & Structural Biology and Principal Investigator within the Division of Biological Chemistry and Drug Discovery. Alessio is the recipient of the 2014 Talented Young Italian award, the 2015 EFMC Prize for Young Medicinal Chemist in Academia, the 2015 ICBS Young Chemical Biologist Award, the 2016 RSC Capps Green Zomaya Award in medicinal computational chemistry, and the 2016 MedChemComm Emerging Investigator Lectureship. Current Research Research in the Ciulli Lab is concerned with understanding molecular recognition of protein-protein interactions (PPIs) within the ubiquitin/proteasome and the chromatin/nucleosomes systems, and exploiting druggability to small molecule modulators for chemical biology and drug discovery. We employ a question-driven, multi-disciplinary approach that combines chemical, biochemical, biophysical and structural techniques with the concepts of fragment-based and structure-based drug design. Current research efforts are directed towards interrogating PPIs and protein recognition within protein families of biological and medical relevance including 1. -

Year in Review
Year in review For the year ended 31 March 2017 Trustees2 Executive Director YEAR IN REVIEW The Trustees of the Society are the members Dr Julie Maxton of its Council, who are elected by and from Registered address the Fellowship. Council is chaired by the 6 – 9 Carlton House Terrace President of the Society. During 2016/17, London SW1Y 5AG the members of Council were as follows: royalsociety.org President Sir Venki Ramakrishnan Registered Charity Number 207043 Treasurer Professor Anthony Cheetham The Royal Society’s Trustees’ report and Physical Secretary financial statements for the year ended Professor Alexander Halliday 31 March 2017 can be found at: Foreign Secretary royalsociety.org/about-us/funding- Professor Richard Catlow** finances/financial-statements Sir Martyn Poliakoff* Biological Secretary Sir John Skehel Members of Council Professor Gillian Bates** Professor Jean Beggs** Professor Andrea Brand* Sir Keith Burnett Professor Eleanor Campbell** Professor Michael Cates* Professor George Efstathiou Professor Brian Foster Professor Russell Foster** Professor Uta Frith Professor Joanna Haigh Dame Wendy Hall* Dr Hermann Hauser Professor Angela McLean* Dame Georgina Mace* Dame Bridget Ogilvie** Dame Carol Robinson** Dame Nancy Rothwell* Professor Stephen Sparks Professor Ian Stewart Dame Janet Thornton Professor Cheryll Tickle Sir Richard Treisman Professor Simon White * Retired 30 November 2016 ** Appointed 30 November 2016 Cover image Dancing with stars by Imre Potyó, Hungary, capturing the courtship dance of the Danube mayfly (Ephoron virgo). YEAR IN REVIEW 3 Contents President’s foreword .................................. 4 Executive Director’s report .............................. 5 Year in review ...................................... 6 Promoting science and its benefits ...................... 7 Recognising excellence in science ......................21 Supporting outstanding science ..................... -

LONG-TERM MEMBERS 25+ Years of Membership
LONG-TERM MEMBERS 25+ Years of Membership Stuart A. Aaronson, MD Stephen P. Ackland, MBBS Carol Aghajanian, MD Steven A. Akman, MD Icahn School of Medicine at Mount Sinai University of Newcastle Memorial Sloan Kettering Cancer Center Roper St. Francis Healthcare United States Australia United States United States Active Member Active Member Active Member Active Member 38 Years of Membership 33 Years of Membership 27 Years of Membership 35 Years of Membership Cory Abate-Shen, PhD Edward M. Acton, PhD Irina U. Agoulnik, PhD Emmanuel T. Akporiaye, PhD Columbia University Irving Medical United States Florida International University Verana Therapeutics Center Emeritus Member United States United States United States 42 Years of Membership Active Member Emeritus Member Active Member 25 Years of Membership 31 Years of Membership 26 Years of Membership David J. Adams, PhD Duke University Medical Center Imran Ahmad, PhD Ala-Eddin Al Moustafa, PhD James L. Abbruzzese, MD United States Northwestern Medicine McGill University Duke University Emeritus Member United States Canada United States 32 Years of Membership Active Member Active Member Active Member 25 Years of Membership 26 Years of Membership 32 Years of Membership Gregory P. Adams, PhD Elucida Oncology Nihal Ahmad, PhD Abdul Al Saadi, PhD Ehtesham A. Abdi, MBBS United States Univ. of Wisconsin Madison Sch. of Med. William Beaumont Hospital The Tweed Hospital Active Member & Public Health United States Australia 29 Years of Membership United States Emeritus Member Emeritus Member Active Member 52 Years of Membership 33 Years of Membership Lucile L. Adams-Campbell, PhD 25 Years of Membership Georgetown Lombardi Comprehensive Suresh K. -

Biographical Sketch Format Page
BIOGRAPHICAL SKETCH NAME POSITION TITLE Professor Paul Workman FRS FMedSci 1. Chief Executive and President, The Institute of Cancer Research (ICR), London UK 2. Director, CRUK-ICR Centre in association with The Royal Marsden NHS Trust, London UK 3. Harrap Professor of Pharmacology and Therapeutics, University of London, UK EDUCATION/TRAINING DEGREE INSTITUTION AND LOCATION (if YY FIELD OF STUDY applicable) University of Leicester, UK BSc (Hons) 1973 Biological Sciences University of Leeds, UK PhD 1977 Cancer Pharmacology A. Personal Statement I am an experienced cancer research scientist with extensive leadership and administrative experience in academia and industry. I am passionate about translating our rapidly emerging understanding of fundamental cancer biology with a sense of urgency into new personalized medicines for cancer patients. I am currently Chief Executive and President of The Institute of Cancer Research (ICR), London and Harrap Professor of Pharmacology and Therapeutics at ICR – a college of the University of London. In addition, I am Director of the CRUK-ICR Centre in association with The Royal Marsden NHS Trust (RM). Furthermore, I am the inaugural Director of the ICR-Imperial College Cancer Research Centre of Excellence in Cancer Research which was established in 2016 to bring together – in synergistic convergence science – Imperial’s strengths in engineering, physical, natural, and data sciences and medicine with ICR’s strengths in basic and applied cancer research, as well as forming the basis for an application to become a CRUK Major Centre, addressing all aspects of cancer prevention, early detection and treatment. ICR came top in REF2014 and ICR/RM has been ranked as one of the top four cancer centres in the world for the impact of its publications (Thomson Reuters Evidence). -

Uril-Based Monodisperse Microcapsules Self- Assembled Within Microfluidic Droplets
Submitted to Accounts of Chemical Research This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies. Cucurbit[n]uril -based Monodisperse Microcapsules Self - Assembled within Microfluidic Droplets: A Versatile Approach for Supramolecular Architectures and Materials Journal: Accounts of Chemical Research Manuscript ID ar-2016-00429g.R2 Manuscript Type: Article Date Submitted by the Author: 22-Nov-2016 Complete List of Authors: Liu, Ji; Melville Laboratory for Polymer Synthesis, Department of Chemistry Lan, Yang; University of Cambridge, Melville Laboratory for Polymer Synthesis, Department of Chemistry Yu, Ziyi; University of Cambridge, Chemistry Tan, Cindy; University of Cambridge, Melville Laboratory for Polymer Synthesis, Department of Chemistry; MARA University of Technology - Sarawak Campus Parker, Richard; University of Cambridge, Chemistry Abell, Chris; University of Cambridge, Chemistry Scherman, Oren; University of Cambridge, Melville Laboratory for Polymer Synthesis, Department of Chemistry ACS Paragon Plus Environment Page 1 of 20 Submitted to Accounts of Chemical Research 1 2 3 4 5 Cucurbit[n]uril-based Microcapsules Self-Assembled within Microfluidic 6 Droplets: A Versatile Approach for Supramolecular Architectures and Materials 7 Ji Liu,† Yang Lan,† Ziyi Yu,∗,‡ Cindy S.Y. Tan,†,¶ Richard M. Parker,‡ Chris Abell,∗,‡ and 8 Oren A. Scherman∗,† 9 † Melville Laboratory for Polymer Synthesis, Department of Chemistry, University of Cambridge, Lensfield 10 Road, Cambridge CB2 1EW, UK. 11 ‡ Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK. 12 ¶Faculty of Applied Sciences, Universiti Teknologi MARA, 94300 Kota Samarahan, Sarawak, Malaysia.