EHD2 Is a Mechanotransducer Connecting Caveolae Dynamics with Gene Transcription
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Membrane Tension Buffering by Caveolae: a Role in Cancer?
Cancer and Metastasis Reviews (2020) 39:505–517 https://doi.org/10.1007/s10555-020-09899-2 Membrane tension buffering by caveolae: a role in cancer? Vibha Singh1 & Christophe Lamaze1 Published online: 30 May 2020 # Springer Science+Business Media, LLC, part of Springer Nature 2020 Abstract Caveolae are bulb-like invaginations made up of two essential structural proteins, caveolin-1 and cavins, which are abundantly present at the plasma membrane of vertebrate cells. Since their discovery more than 60 years ago, the function of caveolae has been mired in controversy. The last decade has seen the characterization of new caveolae components and regulators together with the discovery of additional cellular functions that have shed new light on these enigmatic structures. Early on, caveolae and/ or caveolin-1 have been involved in the regulation of several parameters associated with cancer progression such as cell migration, metastasis, angiogenesis, or cell growth. These studies have revealed that caveolin-1 and more recently cavin-1 have a dual role with either a negative or a positive effect on most of these parameters. The recent discovery that caveolae can act as mechanosensors has sparked an array of new studies that have addressed the mechanobiology of caveolae in various cellular functions. This review summarizes the current knowledge on caveolae and their role in cancer development through their activity in membrane tension buffering. We propose that the role of caveolae in cancer has to be revisited through their response to the mechanical forces encountered by cancer cells during tumor mass development. Keywords Caveolae . Cancer . Mechanosensing . Mechanotransdcution . Membrane tension . -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genomic and Expression Profiling of Chromosome 17 in Breast Cancer Reveals Complex Patterns of Alterations and Novel Candidate Genes
[CANCER RESEARCH 64, 6453–6460, September 15, 2004] Genomic and Expression Profiling of Chromosome 17 in Breast Cancer Reveals Complex Patterns of Alterations and Novel Candidate Genes Be´atrice Orsetti,1 Me´lanie Nugoli,1 Nathalie Cervera,1 Laurence Lasorsa,1 Paul Chuchana,1 Lisa Ursule,1 Catherine Nguyen,2 Richard Redon,3 Stanislas du Manoir,3 Carmen Rodriguez,1 and Charles Theillet1 1Ge´notypes et Phe´notypes Tumoraux, EMI229 INSERM/Universite´ Montpellier I, Montpellier, France; 2ERM 206 INSERM/Universite´ Aix-Marseille 2, Parc Scientifique de Luminy, Marseille cedex, France; and 3IGBMC, U596 INSERM/Universite´Louis Pasteur, Parc d’Innovation, Illkirch cedex, France ABSTRACT 17q12-q21 corresponding to the amplification of ERBB2 and collinear genes, and a large region at 17q23 (5, 6). A number of new candidate Chromosome 17 is severely rearranged in breast cancer. Whereas the oncogenes have been identified, among which GRB7 and TOP2A at short arm undergoes frequent losses, the long arm harbors complex 17q21 or RP6SKB1, TBX2, PPM1D, and MUL at 17q23 have drawn combinations of gains and losses. In this work we present a comprehensive study of quantitative anomalies at chromosome 17 by genomic array- most attention (6–10). Furthermore, DNA microarray studies have comparative genomic hybridization and of associated RNA expression revealed additional candidates, with some located outside current changes by cDNA arrays. We built a genomic array covering the entire regions of gains, thus suggesting the existence of additional amplicons chromosome at an average density of 1 clone per 0.5 Mb, and patterns of on 17q (8, 9). gains and losses were characterized in 30 breast cancer cell lines and 22 Our previous loss of heterozygosity mapping data pointed to the primary tumors. -

Supplemental Material For
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt1,2, Sourav Bandyopadhyay1,3 and Frank McCormick1 Authors Affiliations: 1-Helen Diller Family Comprehensive Cancer Center; 2-Biomedical Sciences Graduate Program, University of California San Francisco, San Francisco, California; 3-California Institute for Quantitative Biosciences, San Francisco, California. Supplemental Figure Legends Table S1. EMT associated changes in gene expression. Significantly (A) up- and (B) down- regulated genes in comparing H358-TwistER cells to control H358-GFP expressing cells treated with 100nM 4OHT for 12 days. The four columns on the right are associated with significantly upregulated genes, while the four columns furthest left described significantly downregulated genes. The first column for the up- and downregulated genes shows the Probe ID associated with each gene from the Affymetric Human Gene 1.0 ST microarrays. The gene name are shown in the next column, followed by the log2(fold change) in expression for each gene after 4OHT treatment in the H358-TwistER cells. The final column shows the p-value for each gene adjusted for the false discovery rate. Figure S1. Akt inhibition reduces serum-independent proliferation. (A) Proliferation of H358- TwistER cells with increasing concentrations (uM) of GSK-690693 (top) or MK-2206 (bottom). (B) Lysates from H358-TwistER cells treated for 48hrs in serum free media with indicated concentrations of GSK-690693 (top), or MK-2206 (bottom). Figure S2. The effects of TGFβ1 are specific to epithelial cells and lead to reduced serum- independent proliferation. (A) Proliferation of untreated (top) or TGFβ1 pre-treated (bottom) H358 and H441 cells in the presence (10%) or absence (SS) of serum. -

Functional Architecture of the Reb1-Ter Complex of PNAS PLUS Schizosaccharomyces Pombe
Functional architecture of the Reb1-Ter complex of PNAS PLUS Schizosaccharomyces pombe Rahul Jaiswala,1,2, Malay Choudhuryb,2, Shamsu Zamanb, Samarendra Singhb,3, Vishaka Santosha, Deepak Bastiab,4, and Carlos R. Escalantea,4 aDepartment of Physiology and Biophysics, Virginia Commonwealth University, Richmond, VA 23298; and bDepartment of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC 29425 Edited by Lucia B. Rothman-Denes, The University of Chicago, Chicago, IL, and approved February 26, 2016 (received for review December 28, 2015) Reb1 of Schizosaccharomyces pombe represents a family of multi- consisting of purified pol I, Reb1 (ScReb1), the Ter site, and an functional proteins that bind to specific terminator sites (Ter) and upstream AT-rich release element were necessary and sufficient cause polar termination of transcription catalyzed by RNA polymer- for transcription termination (7). However, recent work has shown ase I (pol I) and arrest of replication forks approaching the Ter sites that Nsi1 (Ytt1) is also responsible for transcription termination in from the opposite direction. However, it remains to be investigated S. cerevisiae (28). whether the same mechanism causes arrest of both DNA transac- In addition to their aforementioned role, many of these proteins tions. Here, we present the structure of Reb1 as a complex with a are also involved in replication termination and transcription acti- Ter site at a resolution of 2.7 Å. Structure-guided molecular genetic vation. For instance, in S. pombe, the Reb1 (Sp.Reb1) protein binds analyses revealed that it has distinct and well-defined DNA binding to tandem pairs of terminator sites (Ter2 and Ter3) located on the and transcription termination (TTD) domains. -

Stromal CAVIN1 Controls Prostate Cancer Microenvironment and Metastasis by Modulating Lipid Distribution and Inflammatory Signaling
Published OnlineFirst June 3, 2020; DOI: 10.1158/1541-7786.MCR-20-0364 MOLECULAR CANCER RESEARCH | TUMOR MICROENVIRONMENT AND IMMUNOBIOLOGY Stromal CAVIN1 Controls Prostate Cancer Microenvironment and Metastasis by Modulating Lipid Distribution and Inflammatory Signaling Jin-Yih Low1, W. Nathaniel Brennen2, Alan K. Meeker2,3,4, Elina Ikonen5,6, Brian W. Simons7, and Marikki Laiho1,2 ABSTRACT ◥ Lipid uptake occurs through caveolae, plasma membrane invagi- accumulation and increases inflammation. Stromal cells lacking nations formed by caveolins (CAV) and caveolae-associated protein 1 CAVIN1 enhance prostate cancer cell migration and invasion. (CAVIN1). Genetic alterations of CAV1N1 and CAV1 modify lipid Remarkably, they increase lipid uptake and M2 inflammatory mac- metabolism and underpin lipodystrophy syndromes. Lipids contrib- rophage infiltration in the primary tumors and metastasis to distant ute to tumorigenesis by providing fuel to cancer metabolism and sites. Our data support the concept that stromal cells contribute to supporting growth and signaling. Tumor stroma promotes tumor prostate cancer aggressiveness by modulating lipid content and proliferation, invasion, and metastasis, but how stromal lipids influ- inflammation in the tumor microenvironment. ence these processes remain to be defined. Here, we show that stromal CAVIN1 regulates lipid abundance in the prostate cancer microen- Implications: This study showed that stromal CAVIN1 suppresses vironment and suppresses metastasis. We show that depletion of prostate cancer metastasis by modulating tumor microenvironment, CAVIN1 in prostate stromal cells markedly reduces their lipid droplet lipid content, and inflammatory response. Introduction Mice and humans with targeted disruption or mutations of CAVIN1 are affected by numerous abnormalities including lipodystrophy, Caveolae, ultrastructural microdomains at the plasma membrane, muscular dystrophy, cardiovascular disease, and diabetes (8, 13–16). -

Discovery and Systematic Characterization of Risk Variants and Genes For
medRxiv preprint doi: https://doi.org/10.1101/2021.05.24.21257377; this version posted June 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . 1 Discovery and systematic characterization of risk variants and genes for 2 coronary artery disease in over a million participants 3 4 Krishna G Aragam1,2,3,4*, Tao Jiang5*, Anuj Goel6,7*, Stavroula Kanoni8*, Brooke N Wolford9*, 5 Elle M Weeks4, Minxian Wang3,4, George Hindy10, Wei Zhou4,11,12,9, Christopher Grace6,7, 6 Carolina Roselli3, Nicholas A Marston13, Frederick K Kamanu13, Ida Surakka14, Loreto Muñoz 7 Venegas15,16, Paul Sherliker17, Satoshi Koyama18, Kazuyoshi Ishigaki19, Bjørn O Åsvold20,21,22, 8 Michael R Brown23, Ben Brumpton20,21, Paul S de Vries23, Olga Giannakopoulou8, Panagiota 9 Giardoglou24, Daniel F Gudbjartsson25,26, Ulrich Güldener27, Syed M. Ijlal Haider15, Anna 10 Helgadottir25, Maysson Ibrahim28, Adnan Kastrati27,29, Thorsten Kessler27,29, Ling Li27, Lijiang 11 Ma30,31, Thomas Meitinger32,33,29, Sören Mucha15, Matthias Munz15, Federico Murgia28, Jonas B 12 Nielsen34,20, Markus M Nöthen35, Shichao Pang27, Tobias Reinberger15, Gudmar Thorleifsson25, 13 Moritz von Scheidt27,29, Jacob K Ulirsch4,11,36, EPIC-CVD Consortium, Biobank Japan, David O 14 Arnar25,37,38, Deepak S Atri39,3, Noël P Burtt4, Maria C Costanzo4, Jason Flannick40, Rajat M 15 Gupta39,3,4, Kaoru Ito18, Dong-Keun Jang4, -
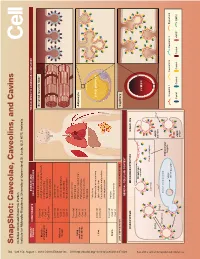
Snapshot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G
704 Cell SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton 154 Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia , August 1,2013©2013 Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.07.009 ORGAN/ COMPONENTS DISEASE AND TISSUE-SPECIFIC CAVEOLAR COMPLEXES TISSUE CELLULAR PROCESS Rippling muscle disease Caveolin3 Striated muscle fiber Limb girdle muscular dystrophy Muscle Cavin1 skeletal and Cardiomyopathy Cavin4 cardiac Mechanoprotection Cav1 (heart) T-tubule formation Caveolin1 Lipodystrophy Caveolin2 Lipotoxicity Adipose Cavin1 Fatty acid regulation tissue Cavin2 Insulin signaling Cavin3 Mechanoprotection Atherosclerosis Caveolin1 Inammation Caveolin2 Lung Pulmonary hypertension and other Cavin1 Adipocyte Pulmonary brosis endothelia Cavin2 Mechanosensation Cavin3 Signaling Fatty liver Hepatocellular carcinoma Caveolin1 LIPID DROPLET Liver Lipid metabolism Caveolin2 Carbohydrate metabolism Liver regeneration Caveolin1 Autism Brain Cavin1 Schizophrenia Cavin3 LOW EXPRESSION HIGH EXPRESSION Capillary GENERAL CELLULAR CONTEXT ENDOCYTOSIS MECHANOPROTECTION SIGNALING See online version for legend and references. and legend for version online See Membrane stretch LUMEN Cavin complex Caveola EHD2 eNOS Ca2+ inactive Rab5 Dynamin2 EARLY ENDOSOME Intracellular targets MVB/ Caveolin 1 Caveolin 2 Caveolin 3 Dynamin2 late endosome eNOS Cavin1 Cavin2 Cavin3 Cavin4 eNOS EHD2 active SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia Caveolae, submicroscopic bulb-shaped plasma membrane pits, are an abundant feature of many mammalian cells (Parton and del Pozo, 2013). Caveolae and the major proteins of caveolae, caveolins (Rothberg et al., 1992), and cavins (Hill et al., 2008), are linked to a number of human diseases such as muscular dystrophy, cardiomyopathy, and lipodys- trophy (see Glossary) (Bruno et al., 1993; Fernández et al., 2006; Hayashi et al., 2009). -

Congenital Generalized Lipodystrophy
Lima JG, Dos Santos MCF, de Melo Campos JTA. J Rare Dis Res Treat. (2018) 3(2): 1-6 Journal of www.rarediseasesjournal.com Rare Diseases Research & Treatment Mini-review Open Access Congenital Generalized Lipodystrophy Josivan Gomes Lima1*, Marcel Catão Ferreira dos Santos1, Julliane Tamara Araújo de Melo Campos2 1Departamento de medicina clínica, disciplina de endocrinologia e metabologia. Hospital Universitário Onofre Lopes, Universidade Federal do Rio Grande do Norte (UFRN), Natal, RN, Brazil. 2Faculty of Health Sciences of Trairi, Federal University of Rio Grande do North (UFRN), Natal, RN, Brazil ABSTRACT Article Info Article Notes Congenital Generalized Lipodystrophy (CGL) is a rare and severe autosomal Received: April 02, 2018 recessive disease. Patients are defective in the storage of body fat and, Accepted: May 11, 2018 consequently, they deposit fat in ectopic tissues, mainly liver, and can develop cirrhosis. Insulin resistance is a typical finding, causing diabetes that require *Correspondence: high daily doses of insulin. In the state of Rio Grande do Norte, Brazil, we Dr. Josivan Gomes Lima, Departamento de medicina clínica, disciplina de endocrinologia e metabologia. Hospital have one of the largest cohorts of patients with CGL. In this article, we review Universitário Onofre Lopes, Universidade Federal do Rio pathophysiology, clinical picture and treatment of this disease. Grande do Norte (UFRN), Natal, RN, Brazil; Email: [email protected] © 2018 Lima JG. This article is distributed under the terms of Introduction the Creative Commons Attribution 4.0 International License. Type 2 diabetes is a world health problem, and usually results Keywords Lipodystrophy from excessive weight and increased visceral fat causing peripheral Berardinelli-Seip syndrome insulin resistance and an inability of the pancreas to release Diabetes insulin to compensate this resistance. -

Congenital Generalized Lipodystrophy
European Journal of Human Genetics (2016) 24, doi:10.1038/ejhg.2016.53 & 2016 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 1018-4813/16 www.nature.com/ejhg CLINICAL UTILITY GENE CARD Clinical Utility Gene Card for: Congenital Generalized Lipodystrophy Isabelle Jéru*,1,2,3, Camille Vatier2,3,4, David Araujo-Vilar5, Corinne Vigouroux1,2,3,4 and Olivier Lascols1,2,3 European Journal of Human Genetics (2016) 24, doi:10.1038/ejhg.2016.53; published online 18 May 2016 1. DISEASE CHARACTERISTICS molecular defects have been reported: nonsense, missense, splice-site 1.1 Name of the disease (synonyms) variants, deletions and insertions.3 Most of them result in frameshift There are four subclasses of Congenital Generalized Lipodystrophy and/or truncated proteins, which are likely to lead to the complete loss (CGL), also named Berardinelli–Seip Congenital Lipodystrophy (BSCL): of AGPAT2 function. This can be demonstrated in vitro by measure- Type 1 CGL (CGL1). ment of AGPAT2 enzymatic activity.4 Type 2 CGL (CGL2). BSCL2—CGL2 is an autosomal recessive disorder. Several dozen Type 3 CGL (CGL3). disease-causing variants have been described. The following molecular Type 4 CGL (CGL4). defects have been reported: nonsense, missense, splice-site variants, It is important to underline that CGL is an expanding group of deletions and insertions.3,5 Most of them result in frameshift and/or fi disorders, whose classi cation is still underway. In addition to the truncated proteins, which are likely to lead to complete loss of BSCL2 diseases known as CGL in OMIM, generalized lipodystrophies can be function (for more details, see http://databases.lovd.nl/shared/variants/ encountered in other Mendelian disorders including severe insulin- BSCL2/unique). -

Caveolin-1 Expression and Cavin Stability Regulate Caveolae Dynamics in Adipocyte Lipid Store Fluctuation
4032 Diabetes Volume 63, December 2014 Nolwenn Briand,1 Cécilia Prado,1 Guillaume Mabilleau,2 Françoise Lasnier,1 Xavier Le Lièpvre,1 Jeffrey D. Covington,3 Eric Ravussin,3 Soazig Le Lay,4 and Isabelle Dugail5 Caveolin-1 Expression and Cavin Stability Regulate Caveolae Dynamics in Adipocyte Lipid Store Fluctuation Diabetes 2014;63:4032–4044 | DOI: 10.2337/db13-1961 Adipocytes specialized in the storage of energy as fat Caveolae are small flask-shaped invaginations of the plasma are among the most caveolae-enriched cell types. Loss membrane (1) that are found with remarkable abundance in of caveolae produces lipodystrophic diabetes in humans, endothelial cells, myotubes, and adipocytes. They are consid- which cannot be reversed by endothelial rescue of ered a subset of the so-called lipid raft domains and segregate caveolin expression in mice, indicating major impor- a number of membrane-related processes (2). An accepted tance of adipocyte caveolae. However, how caveolae paradigm is that caveolae formation is primarily driven by participate in fat cell functions is poorly understood. the assembly of a cytoplasmic coat consisting of oligomeric We investigated dynamic conditions of lipid store caveolins (3), a protein family with 3 highly-related members fluctuations and demonstrate reciprocal regulation of (caveolin-1 through -3). Invalidation of individual caveolin caveolae density and fat cell lipid droplet storage. We genes led to the generation of mice models lacking caveolae METABOLISM identified caveolin-1 expression as a crucial step in in all cell types or in a tissue-restricted manner (4). Caveolin- adipose cell lines and in mice to raise the density of 1–null mice, which also lack caveolin-2, suffer from severe caveolae, to increase adipocyte ability to accommodate vascular dysfunction and pulmonary defects (5,6) and de- larger lipid droplets, and to promote cell expansion velop lipodystrophy (7), a metabolic phenotype that cannot by increased glucose utilization. -

Transcription Termination by Nuclear RNA Polymerases
Downloaded from genesdev.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW Transcription termination by nuclear RNA polymerases Patricia Richard and James L. Manley1 Department of Biological Sciences, Columbia University, New York, New York 10027, USA Gene transcription in the cell nucleus is a complex and few base pairs to several kilobases downstream from the highly regulated process. Transcription in eukaryotes 39-end of the mature RNA (Proudfoot 1989). RNAPII requires three distinct RNA polymerases, each of which transcription termination is coupled to 39-end processing employs its own mechanisms for initiation, elongation, of the pre-mRNA (Birse et al. 1998; Hirose and Manley and termination. Termination mechanisms vary consid- 2000; Yonaha and Proudfoot 2000; Proudfoot 2004; erably, ranging from relatively simple to exceptionally Buratowski 2005), and an intact polyadenylation signal complex. In this review, we describe the present state of has long been known to be necessary for transcription knowledge on how each of the three RNA polymerases termination of protein-coding genes in human and yeast terminates and how mechanisms are conserved, or vary, cells (Whitelaw and Proudfoot 1986; Logan et al. 1987; from yeast to human. Connelly and Manley 1988). RNAPIII and RNAPI termination appear simpler than Transcription in eukaryotes is performed by three RNA RNAPII termination. RNAPIII terminates transcription polymerases, which are functionally and structurally at T-rich sequences located a short distance from the related (Cramer et al. 2008). RNA polymerase II (RNAPII) mature RNA 39-end and seems to involve at most a is responsible for transcription of protein-coding genes limited number of auxiliary factors (Cozzarelli et al.