Bmerb Domains Are Bivalent Rab8 Family Effectors Evolved by Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![MICAL1 Mouse Monoclonal Antibody [Clone ID: OTI6G2] Product Data](https://docslib.b-cdn.net/cover/9450/mical1-mouse-monoclonal-antibody-clone-id-oti6g2-product-data-39450.webp)
MICAL1 Mouse Monoclonal Antibody [Clone ID: OTI6G2] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA501844 MICAL1 Mouse Monoclonal Antibody [Clone ID: OTI6G2] Product data: Product Type: Primary Antibodies Clone Name: OTI6G2 Applications: FC, IF, WB Recommended Dilution: WB 1:500, IF 1:100, FLOW 1:100 Reactivity: Human Host: Mouse Isotype: IgG1 Clonality: Monoclonal Immunogen: Full length human recombinant protein of human MICAL1 (NP_073602) produced in HEK293T cell. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 1 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 117.7 kDa Gene Name: microtubule associated monooxygenase, calponin and LIM domain containing 1 Database Link: NP_073602 Entrez Gene 64780 Human Q8TDZ2 Background: May be a cytoskeletal regulator that connects NEDD9 to intermediate filaments Synonyms: MICAL; MICAL-1; NICAL This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 MICAL1 Mouse Monoclonal Antibody [Clone ID: OTI6G2] – TA501844 Product images: HEK293T cells were transfected with the pCMV6- ENTRY control (Left lane) or pCMV6-ENTRY MICAL1 ([RC208308], Right lane) cDNA for 48 hrs and lysed. Equivalent amounts of cell lysates (5 ug per lane) were separated by SDS-PAGE and immunoblotted with anti-MICAL1. -

Membrane Tension Buffering by Caveolae: a Role in Cancer?
Cancer and Metastasis Reviews (2020) 39:505–517 https://doi.org/10.1007/s10555-020-09899-2 Membrane tension buffering by caveolae: a role in cancer? Vibha Singh1 & Christophe Lamaze1 Published online: 30 May 2020 # Springer Science+Business Media, LLC, part of Springer Nature 2020 Abstract Caveolae are bulb-like invaginations made up of two essential structural proteins, caveolin-1 and cavins, which are abundantly present at the plasma membrane of vertebrate cells. Since their discovery more than 60 years ago, the function of caveolae has been mired in controversy. The last decade has seen the characterization of new caveolae components and regulators together with the discovery of additional cellular functions that have shed new light on these enigmatic structures. Early on, caveolae and/ or caveolin-1 have been involved in the regulation of several parameters associated with cancer progression such as cell migration, metastasis, angiogenesis, or cell growth. These studies have revealed that caveolin-1 and more recently cavin-1 have a dual role with either a negative or a positive effect on most of these parameters. The recent discovery that caveolae can act as mechanosensors has sparked an array of new studies that have addressed the mechanobiology of caveolae in various cellular functions. This review summarizes the current knowledge on caveolae and their role in cancer development through their activity in membrane tension buffering. We propose that the role of caveolae in cancer has to be revisited through their response to the mechanical forces encountered by cancer cells during tumor mass development. Keywords Caveolae . Cancer . Mechanosensing . Mechanotransdcution . Membrane tension . -

EHD2 Is a Mechanotransducer Connecting Caveolae Dynamics with Gene Transcription
EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription Satish Kailasam Mani, Cesar Valades-Cruz, Stéphanie Torrino, Wei-Wei Shen, Cedric Blouin, Satish Kailasam Mani, Christine Viaris de Lesegno, Pierre Bost, Alexandre Grassart, Darius Köster, et al. To cite this version: Satish Kailasam Mani, Cesar Valades-Cruz, Stéphanie Torrino, Wei-Wei Shen, Cedric Blouin, et al.. EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription. Journal of Cell Biology, Rockefeller University Press, 2018, 217 (12), pp.4092-4105. 10.1083/jcb.201801122. inserm- 02426440 HAL Id: inserm-02426440 https://www.hal.inserm.fr/inserm-02426440 Submitted on 2 Jan 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License REPORT EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription Stéphanie Torrino1,2,3*, Wei‑Wei Shen1,2,3*, Cédric M. Blouin1,2,3, Satish Kailasam Mani1,2,3, Christine Viaris de Lesegno1,2,3, Pierre Bost4,5, Alexandre Grassart6, Darius Köster7, Cesar Augusto Valades‑Cruz2,3,8, Valérie Chambon2,3,8, Ludger Johannes2,3,8, Paolo Pierobon9, Vassili Soumelis4, Catherine Coirault10, Stéphane Vassilopoulos10, and Christophe Lamaze1,2,3 Caveolae are small invaginated pits that function as dynamic mechanosensors to buffer tension variations at the plasma membrane. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplemental Material For
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt1,2, Sourav Bandyopadhyay1,3 and Frank McCormick1 Authors Affiliations: 1-Helen Diller Family Comprehensive Cancer Center; 2-Biomedical Sciences Graduate Program, University of California San Francisco, San Francisco, California; 3-California Institute for Quantitative Biosciences, San Francisco, California. Supplemental Figure Legends Table S1. EMT associated changes in gene expression. Significantly (A) up- and (B) down- regulated genes in comparing H358-TwistER cells to control H358-GFP expressing cells treated with 100nM 4OHT for 12 days. The four columns on the right are associated with significantly upregulated genes, while the four columns furthest left described significantly downregulated genes. The first column for the up- and downregulated genes shows the Probe ID associated with each gene from the Affymetric Human Gene 1.0 ST microarrays. The gene name are shown in the next column, followed by the log2(fold change) in expression for each gene after 4OHT treatment in the H358-TwistER cells. The final column shows the p-value for each gene adjusted for the false discovery rate. Figure S1. Akt inhibition reduces serum-independent proliferation. (A) Proliferation of H358- TwistER cells with increasing concentrations (uM) of GSK-690693 (top) or MK-2206 (bottom). (B) Lysates from H358-TwistER cells treated for 48hrs in serum free media with indicated concentrations of GSK-690693 (top), or MK-2206 (bottom). Figure S2. The effects of TGFβ1 are specific to epithelial cells and lead to reduced serum- independent proliferation. (A) Proliferation of untreated (top) or TGFβ1 pre-treated (bottom) H358 and H441 cells in the presence (10%) or absence (SS) of serum. -

Pan-Cancer Analysis Identifies Mutations in SUGP1 That Recapitulate Mutant SF3B1 Splicing Dysregulation
Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation Zhaoqi Liua,b,c,1, Jian Zhangd,1, Yiwei Suna,c, Tomin E. Perea-Chambleea,b,c, James L. Manleyd,2, and Raul Rabadana,b,c,2 aProgram for Mathematical Genomics, Columbia University, New York, NY 10032; bDepartment of Systems Biology, Columbia University, New York, NY 10032; cDepartment of Biomedical Informatics, Columbia University, New York, NY 10032; and dDepartment of Biological Sciences, Columbia University, New York, NY 10027 Contributed by James L. Manley, March 2, 2020 (sent for review January 2, 2020; reviewed by Kristen Lynch and Gene Yeo) The gene encoding the core spliceosomal protein SF3B1 is the most also resulted in the same splicing defects observed in SF3B1 frequently mutated gene encoding a splicing factor in a variety of mutant cells (11). hematologic malignancies and solid tumors. SF3B1 mutations in- In addition to SF3B1, other SF-encoding genes have also been duce use of cryptic 3′ splice sites (3′ss), and these splicing errors found to be mutated in hematologic malignancies, e.g., U2AF1, contribute to tumorigenesis. However, it is unclear how wide- SRSF2, and ZRSR2. However, these SF gene mutations do not spread this type of cryptic 3′ss usage is in cancers and what is share common alterations in splicing (1, 3), suggesting that dif- the full spectrum of genetic mutations that cause such missplicing. ferent splicing patterns may contribute to different phenotypes To address this issue, we performed an unbiased pan-cancer anal- of cancers. Because SF3B1 is the most frequently mutated ysis to identify genetic alterations that lead to the same aberrant splicing gene, the splicing defects caused by mutant SF3B1 may SF3B1 splicing as observed with mutations. -

The Genetics of Bipolar Disorder
Molecular Psychiatry (2008) 13, 742–771 & 2008 Nature Publishing Group All rights reserved 1359-4184/08 $30.00 www.nature.com/mp FEATURE REVIEW The genetics of bipolar disorder: genome ‘hot regions,’ genes, new potential candidates and future directions A Serretti and L Mandelli Institute of Psychiatry, University of Bologna, Bologna, Italy Bipolar disorder (BP) is a complex disorder caused by a number of liability genes interacting with the environment. In recent years, a large number of linkage and association studies have been conducted producing an extremely large number of findings often not replicated or partially replicated. Further, results from linkage and association studies are not always easily comparable. Unfortunately, at present a comprehensive coverage of available evidence is still lacking. In the present paper, we summarized results obtained from both linkage and association studies in BP. Further, we indicated new potential interesting genes, located in genome ‘hot regions’ for BP and being expressed in the brain. We reviewed published studies on the subject till December 2007. We precisely localized regions where positive linkage has been found, by the NCBI Map viewer (http://www.ncbi.nlm.nih.gov/mapview/); further, we identified genes located in interesting areas and expressed in the brain, by the Entrez gene, Unigene databases (http://www.ncbi.nlm.nih.gov/entrez/) and Human Protein Reference Database (http://www.hprd.org); these genes could be of interest in future investigations. The review of association studies gave interesting results, as a number of genes seem to be definitively involved in BP, such as SLC6A4, TPH2, DRD4, SLC6A3, DAOA, DTNBP1, NRG1, DISC1 and BDNF. -

Rapid Diagnosis of Medulloblastoma Molecular Subgroups
Published OnlineFirst February 16, 2011; DOI: 10.1158/1078-0432.CCR-10-2210 Clinical Cancer Imaging, Diagnosis, Prognosis Research Rapid Diagnosis of Medulloblastoma Molecular Subgroups Ed C. Schwalbe1, Janet C. Lindsey1, Debbie Straughton1, Twala L. Hogg2, Michael Cole1, Hisham Megahed1, Sarra L. Ryan1, Meryl E. Lusher1, Michael D. Taylor4, Richard J. Gilbertson2, David W. Ellison3, Simon Bailey1, and Steven C. Clifford1 Abstract Purpose: Microarray studies indicate medulloblastoma comprises distinct molecular disease subgroups, which offer potential for improved clinical management. Experimental Design: Minimal mRNA expression signatures diagnostic for the Wnt/Wingless (WNT) and Sonic Hedgehog (SHH) subgroups were developed, validated, and used to assign subgroup affiliation in 173 tumors from four independent cohorts, alongside a systematic investigation of subgroup clinical and molecular characteristics. Results: WNT tumors [12% (21/173)] were diagnosed >5 years of age (peak, 10 years), displayed classic histology, CTNNB1 mutation (19/20), and associated chromosome 6 loss, and have previously been associated with favorable prognosis. SHH cases [24% (42/173)] predominated in infants (<3 years) and showed an age-dependent relationship to desmoplastic/nodular pathology; all infant desmoplastic/ nodular cases (previously associated with a good outcome) were SHH-positive, but these relationships broke down in noninfants. PTCH1 mutations were common [34% (11/32)], but PTCH1 exon1c hypermethylation, chromosome 9q and REN (KCTD11) genetic loss were not SHH associated, and SMO or SUFU mutation, PTCH1 exon1a or SUFU hypermethylation did not play a role, indicating novel activating mechanisms in the majority of SHH cases. SHH tumors were associated with an absence of COL1A2 methylation. WNT/SHH-independent medulloblastomas [64% (110/173)] showed all histolo- gies, peaked at 3 and 6 years, and were exclusively associated with chromosome 17p loss. -

Genomic and Transcriptome Analysis Revealing an Oncogenic Functional Module in Meningiomas
Neurosurg Focus 35 (6):E3, 2013 ©AANS, 2013 Genomic and transcriptome analysis revealing an oncogenic functional module in meningiomas XIAO CHANG, PH.D.,1 LINGLING SHI, PH.D.,2 FAN GAO, PH.D.,1 JONATHAN RUssIN, M.D.,3 LIYUN ZENG, PH.D.,1 SHUHAN HE, B.S.,3 THOMAS C. CHEN, M.D.,3 STEVEN L. GIANNOTTA, M.D.,3 DANIEL J. WEISENBERGER, PH.D.,4 GAbrIEL ZADA, M.D.,3 KAI WANG, PH.D.,1,5,6 AND WIllIAM J. MAck, M.D.1,3 1Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California; 2GHM Institute of CNS Regeneration, Jinan University, Guangzhou, China; 3Department of Neurosurgery, Keck School of Medicine, University of Southern California, Los Angeles, California; 4USC Epigenome Center, Keck School of Medicine, University of Southern California, Los Angeles, California; 5Department of Psychiatry, Keck School of Medicine, University of Southern California, Los Angeles, California; and 6Division of Bioinformatics, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California Object. Meningiomas are among the most common primary adult brain tumors. Although typically benign, roughly 2%–5% display malignant pathological features. The key molecular pathways involved in malignant trans- formation remain to be determined. Methods. Illumina expression microarrays were used to assess gene expression levels, and Illumina single- nucleotide polymorphism arrays were used to identify copy number variants in benign, atypical, and malignant me- ningiomas (19 tumors, including 4 malignant ones). The authors also reanalyzed 2 expression data sets generated on Affymetrix microarrays (n = 68, including 6 malignant ones; n = 56, including 3 malignant ones). -
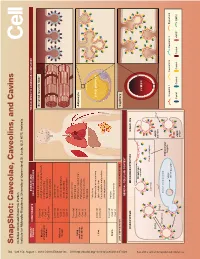
Snapshot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G
704 Cell SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton 154 Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia , August 1,2013©2013 Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.07.009 ORGAN/ COMPONENTS DISEASE AND TISSUE-SPECIFIC CAVEOLAR COMPLEXES TISSUE CELLULAR PROCESS Rippling muscle disease Caveolin3 Striated muscle fiber Limb girdle muscular dystrophy Muscle Cavin1 skeletal and Cardiomyopathy Cavin4 cardiac Mechanoprotection Cav1 (heart) T-tubule formation Caveolin1 Lipodystrophy Caveolin2 Lipotoxicity Adipose Cavin1 Fatty acid regulation tissue Cavin2 Insulin signaling Cavin3 Mechanoprotection Atherosclerosis Caveolin1 Inammation Caveolin2 Lung Pulmonary hypertension and other Cavin1 Adipocyte Pulmonary brosis endothelia Cavin2 Mechanosensation Cavin3 Signaling Fatty liver Hepatocellular carcinoma Caveolin1 LIPID DROPLET Liver Lipid metabolism Caveolin2 Carbohydrate metabolism Liver regeneration Caveolin1 Autism Brain Cavin1 Schizophrenia Cavin3 LOW EXPRESSION HIGH EXPRESSION Capillary GENERAL CELLULAR CONTEXT ENDOCYTOSIS MECHANOPROTECTION SIGNALING See online version for legend and references. and legend for version online See Membrane stretch LUMEN Cavin complex Caveola EHD2 eNOS Ca2+ inactive Rab5 Dynamin2 EARLY ENDOSOME Intracellular targets MVB/ Caveolin 1 Caveolin 2 Caveolin 3 Dynamin2 late endosome eNOS Cavin1 Cavin2 Cavin3 Cavin4 eNOS EHD2 active SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia Caveolae, submicroscopic bulb-shaped plasma membrane pits, are an abundant feature of many mammalian cells (Parton and del Pozo, 2013). Caveolae and the major proteins of caveolae, caveolins (Rothberg et al., 1992), and cavins (Hill et al., 2008), are linked to a number of human diseases such as muscular dystrophy, cardiomyopathy, and lipodys- trophy (see Glossary) (Bruno et al., 1993; Fernández et al., 2006; Hayashi et al., 2009). -

The Human Gene Connectome As a Map of Short Cuts for Morbid Allele Discovery
The human gene connectome as a map of short cuts for morbid allele discovery Yuval Itana,1, Shen-Ying Zhanga,b, Guillaume Vogta,b, Avinash Abhyankara, Melina Hermana, Patrick Nitschkec, Dror Friedd, Lluis Quintana-Murcie, Laurent Abela,b, and Jean-Laurent Casanovaa,b,f aSt. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY 10065; bLaboratory of Human Genetics of Infectious Diseases, Necker Branch, Paris Descartes University, Institut National de la Santé et de la Recherche Médicale U980, Necker Medical School, 75015 Paris, France; cPlateforme Bioinformatique, Université Paris Descartes, 75116 Paris, France; dDepartment of Computer Science, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel; eUnit of Human Evolutionary Genetics, Centre National de la Recherche Scientifique, Unité de Recherche Associée 3012, Institut Pasteur, F-75015 Paris, France; and fPediatric Immunology-Hematology Unit, Necker Hospital for Sick Children, 75015 Paris, France Edited* by Bruce Beutler, University of Texas Southwestern Medical Center, Dallas, TX, and approved February 15, 2013 (received for review October 19, 2012) High-throughput genomic data reveal thousands of gene variants to detect a single mutated gene, with the other polymorphic genes per patient, and it is often difficult to determine which of these being of less interest. This goes some way to explaining why, variants underlies disease in a given individual. However, at the despite the abundance of NGS data, the discovery of disease- population level, there may be some degree of phenotypic homo- causing alleles from such data remains somewhat limited. geneity, with alterations of specific physiological pathways under- We developed the human gene connectome (HGC) to over- come this problem. -

MICAL1 Regulates Actin Cytoskeleton Organization, Directional Cell Migration and the Growth of Human Breast Cancer Cells As Orthotopic Xenograft Tumours
Cancer Letters 519 (2021) 226–236 Contents lists available at ScienceDirect Cancer Letters journal homepage: www.elsevier.com/locate/canlet MICAL1 regulates actin cytoskeleton organization, directional cell migration and the growth of human breast cancer cells as orthotopic xenograft tumours David J. McGarry a, Garett Armstrong a, Giovanni Castino a, Susan Mason b, William Clark b, Robin Shaw b, Lynn McGarry b, Karen Blyth b,c, Michael F. Olson a,d,* a Department of Chemistry and Biology, Ryerson University, Toronto, ON, Canada b Cancer Research UK Beatson Institute, Glasgow, UK c Institute of Cancer Sciences, University of Glasgow, Glasgow, UK d Department of Pharmacology and Toxicology, University of Toronto, Toronto, ON, Canada ARTICLE INFO ABSTRACT Keywords: The Molecule Interacting with CasL 1 (MICAL1) monooxygenase has emerged as an important regulator of Cytoskeleton cytoskeleton organization via actin oxidation. Although filamentous actin (F-actin) increases MICAL1 mono Reactive oxygen species oxygenase activity, hydrogen peroxide (H2O2) is also generated in the absence of F-actin, suggesting that cell morphology diffusible H2O2 might have additional functions. MICAL1 gene disruption by CRISPR/Cas9 in MDA MB 231 cell motility human breast cancer cells knocked out (KO) protein expression, which affected F-actin organization, cell size and cell size Transcription motility. Transcriptomic profilingrevealed that MICAL1 deletion significantlyaffected the expression of over 700 Breast cancer genes, with the majority being reduced in their expression levels. In addition, the absolute magnitudes of reduced MICAL1 gene expression were significantly greater than the magnitudes of increased gene expression. Gene set enrich ment analysis (GSEA) identifiedreceptor regulator activity as the most significantnegatively enriched molecular function gene set.