Membrane Tension Buffering by Caveolae: a Role in Cancer?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

EHD2 Is a Mechanotransducer Connecting Caveolae Dynamics with Gene Transcription
EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription Satish Kailasam Mani, Cesar Valades-Cruz, Stéphanie Torrino, Wei-Wei Shen, Cedric Blouin, Satish Kailasam Mani, Christine Viaris de Lesegno, Pierre Bost, Alexandre Grassart, Darius Köster, et al. To cite this version: Satish Kailasam Mani, Cesar Valades-Cruz, Stéphanie Torrino, Wei-Wei Shen, Cedric Blouin, et al.. EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription. Journal of Cell Biology, Rockefeller University Press, 2018, 217 (12), pp.4092-4105. 10.1083/jcb.201801122. inserm- 02426440 HAL Id: inserm-02426440 https://www.hal.inserm.fr/inserm-02426440 Submitted on 2 Jan 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License REPORT EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription Stéphanie Torrino1,2,3*, Wei‑Wei Shen1,2,3*, Cédric M. Blouin1,2,3, Satish Kailasam Mani1,2,3, Christine Viaris de Lesegno1,2,3, Pierre Bost4,5, Alexandre Grassart6, Darius Köster7, Cesar Augusto Valades‑Cruz2,3,8, Valérie Chambon2,3,8, Ludger Johannes2,3,8, Paolo Pierobon9, Vassili Soumelis4, Catherine Coirault10, Stéphane Vassilopoulos10, and Christophe Lamaze1,2,3 Caveolae are small invaginated pits that function as dynamic mechanosensors to buffer tension variations at the plasma membrane. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplemental Material For
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt1,2, Sourav Bandyopadhyay1,3 and Frank McCormick1 Authors Affiliations: 1-Helen Diller Family Comprehensive Cancer Center; 2-Biomedical Sciences Graduate Program, University of California San Francisco, San Francisco, California; 3-California Institute for Quantitative Biosciences, San Francisco, California. Supplemental Figure Legends Table S1. EMT associated changes in gene expression. Significantly (A) up- and (B) down- regulated genes in comparing H358-TwistER cells to control H358-GFP expressing cells treated with 100nM 4OHT for 12 days. The four columns on the right are associated with significantly upregulated genes, while the four columns furthest left described significantly downregulated genes. The first column for the up- and downregulated genes shows the Probe ID associated with each gene from the Affymetric Human Gene 1.0 ST microarrays. The gene name are shown in the next column, followed by the log2(fold change) in expression for each gene after 4OHT treatment in the H358-TwistER cells. The final column shows the p-value for each gene adjusted for the false discovery rate. Figure S1. Akt inhibition reduces serum-independent proliferation. (A) Proliferation of H358- TwistER cells with increasing concentrations (uM) of GSK-690693 (top) or MK-2206 (bottom). (B) Lysates from H358-TwistER cells treated for 48hrs in serum free media with indicated concentrations of GSK-690693 (top), or MK-2206 (bottom). Figure S2. The effects of TGFβ1 are specific to epithelial cells and lead to reduced serum- independent proliferation. (A) Proliferation of untreated (top) or TGFβ1 pre-treated (bottom) H358 and H441 cells in the presence (10%) or absence (SS) of serum. -

Distinct EH Domains of the Endocytic TPLATE Complex Confer Lipid and Protein Binding
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.29.122911; this version posted May 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding. Klaas Yperman1,2, Anna C. Papageorgiou3,*, Romain Merceron4,5,*, Steven De Munck4,5, Yehudi Bloch4,5, Dominique Eeckhout1,2, Pieter Tack6, Thomas Evangelidis3, Jelle Van Leene1,2, Laszlo Vincze6, Peter Vandenabeele6,7, Martin Potocký8, Geert De Jaeger1,2, Savvas N. Savvides4,5, Konstantinos Tripsianes3, Roman Pleskot1,2,# & Daniel Van Damme1,2,# * Equal contribution # joint senior authorship 1Ghent University, Department of Plant Biotechnology and Bioinformatics, Technologiepark 71, 9052 Ghent, Belgium. 2VIB Center for Plant Systems Biology, Technologiepark 71, 9052 Ghent, Belgium. 3CEITEC-Central European Institute of Technology, Masaryk University, Kamenice 5, 62500, Brno, Czech Republic. 4Department of Biochemistry and Microbiology, Ghent University, 9052 Ghent, Belgium. 5VIB Center for Inflammation Research, 9052 Ghent, Belgium. 6Department of Analytical Chemistry, Ghent University, 9000 Ghent, Belgium 7Archaeometry Research Group, Department of Archaeology, Ghent University, Sint- Pietersnieuwstraat 35, B-9000 Ghent, Belgium 8Institute of Experimental Botany, Academy of Sciences of the Czech Republic, Rozvojová 263, 16502 Prague 6, Czech Republic Abstract Clathrin-mediated endocytosis (CME) is the gatekeeper of the plasma membrane. In contrast to animals and yeasts, CME in plants depends on the TPLATE complex (TPC), an evolutionary ancient adaptor complex. The mechanistic contribution of the individual TPC subunits to plant CME remains however elusive. -

Sphingosine-1-Phosphate Receptor 2 Controls Podosome Components Induced by RANKL Affecting Osteoclastogenesis and Bone Resorption
cells Article Sphingosine-1-Phosphate Receptor 2 Controls Podosome Components Induced by RANKL Affecting Osteoclastogenesis and Bone Resorption Li-Chien Hsu 1, Sakamuri V. Reddy 2, Özlem Yilmaz 1 and Hong Yu 1,* 1 Department of Oral Health Sciences, College of Dental Medicine, Medical University of South Carolina, Charleston, SC 29425, USA; [email protected] (L.-C.H.); [email protected] (Ö.Y.) 2 Department of Pediatrics, Osteoclast Center, Darby Children’s Research Institute, Medical University of South Carolina, Charleston, SC 29425, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-843-792-0635 Received: 17 November 2018; Accepted: 26 December 2018; Published: 1 January 2019 Abstract: Proinflammatory cytokine production, cell chemotaxis, and osteoclastogenesis can lead to inflammatory bone loss. Previously, we showed that sphingosine-1-phosphate receptor 2 (S1PR2), a G protein coupled receptor, regulates inflammatory cytokine production and osteoclastogenesis. However, the signaling pathways regulated by S1PR2 in modulating inflammatory bone loss have not been elucidated. Herein, we demonstrated that inhibition of S1PR2 by a specific S1PR2 antagonist (JTE013) suppressed phosphoinositide 3-kinase (PI3K), mitogen-activated protein kinases (MAPKs), and nuclear factor kappa-B (NF-κB) induced by an oral bacterial pathogen, Aggregatibacter actinomycetemcomitans, and inhibited the release of IL-1β, IL-6, TNF-α, and S1P in murine bone marrow cells. In addition, shRNA knockdown of S1PR2 or treatment by JTE013 suppressed cell chemotaxis induced by bacteria-stimulated cell culture media. Furthermore, JTE013 suppressed osteoclastogenesis and bone resorption induced by RANKL in murine bone marrow cultures. ShRNA knockdown of S1PR2 or inhibition of S1PR2 by JTE013 suppressed podosome components, including PI3K, Src, Pyk2, integrin β3, filamentous actin (F-actin), and paxillin levels induced by RANKL in murine bone marrow cells. -

A Novel Role for Calpain 4 in Podosome Assembly
A NOVEL ROLE FOR CALPAIN 4 IN PODOSOME ASSEMBLY by Thomas Riley Dowler A thesis submitted to the Department of Biochemistry In conformity with the requirements for the degree of Masters of Science Queen’s University Kingston, Ontario, Canada September, 2008 Copyright © Thomas Riley Dowler, 2008 Abstract Podosomes are adhesive and invasive structures which may play an important role in numerous physiological and pathological conditions including angiogenesis, atherosclerosis, and cancer metastasis. Recently, the cysteine protease m-calpain (m- Capn) has been shown to cleave cortactin, an integral component of the podosomal F- actin core, as well as various proteins found in the peripheral adhesive region leading to the disassembly of these dynamic structures. In this study, I investigated whether Capn plays a role in the formation of podosomes downstream of c-Src. I show that: 1) phorbol- 12, 13-dibutyrate (PDBu) as well as c-Src-Y527F expression induces podosome formation in mouse embryonic fibroblasts; 2) PDBu- and constitutively active c-Src- induced podosome formation is inhibited by the knockout of the m- and µ-Capn small regulatory subunit Capn4 in mouse embryonic fibroblasts (Capn4-/-), but is partially restored by re-expression of Capn4; 3) Capn4 localizes to podosomes; and 4) Inhibition of m- and µ-Capn proteolytic activity by the cell permeable calpain inhibitors has little effect on the formation of podosomes downstream of active c-Src. I conclude that Capn4 may play a role in the assembly phase of podosomes independent of calpain proteolytic activity. Work done in collaboration to determine a possible mechanism of action for the role of Capn4 in podosome assembly indicates that a possible binding partner of Capn4, β-PIX, co-localizes with, and shows in vivo association with Capn4. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Poji: a Fiji-Based Tool for Analysis of Podosomes and Associated Proteins Robert Herzog1, Koen Van Den Dries2, Pasquale Cervero1 and Stefan Linder1,*
© 2020. Published by The Company of Biologists Ltd | Journal of Cell Science (2020) 133, jcs238964. doi:10.1242/jcs.238964 TOOLS AND RESOURCES Poji: a Fiji-based tool for analysis of podosomes and associated proteins Robert Herzog1, Koen van den Dries2, Pasquale Cervero1 and Stefan Linder1,* ABSTRACT structure that contains actomyosin contractility regulators such as Podosomes are actin-based adhesion and invasion structures in a lymphocyte-specific protein 1 (LSP1) (Cervero et al., 2018) and variety of cell types, with podosome-forming cells displaying up to supervillin (Bhuwania et al., 2012). Podosomes display a remarkably several hundreds of these structures. Podosome number, distribution broad repertoire of functions, ranging from cell-matrix adhesion and and composition can be affected by experimental treatments or during extracellular matrix degradation, to antigen presentation and regular turnover, necessitating a tool that is able to detect even subtle mechanosensing, and we refer to several reviews that cover these differences in podosomal properties. Here, we present a Fiji-based various aspects (Albiges-Rizo et al., 2009; Alonso et al., 2019; macro code termed ‘Poji’ (‘podosome analysis by Fiji’), which serves as Destaing et al., 2011; Linder, 2007; Linder and Wiesner, 2015; an easy-to-use tool to characterize a variety of cellular and podosomal Paterson and Courtneidge, 2018; van den Dries et al., 2014, 2019). parameters, including area, fluorescence intensity, relative enrichment Of note, especially myeloid cells such as macrophages often form of associated proteins and radial podosome intensity profiles. This tool >500 podosomes per cell, making podosomes a prominent should be useful to gain more detailed insight into the regulation, component of the actin cytoskeleton of these cells. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

In This Issue
By Jane Bradbury In this issue Samp1 makes nuclear moves FOXO3, ROS and apoptosis Diagrams of eukaryotic cells often show the nucleus in the Forkhead box O (FOXO) transcription factors induce centre of the cell, but the position of the nucleus actually apoptosis and regulate the production of reactive oxygen changes during certain developmental stages and cellular species (ROS). In neuronal tumour cells derived from events. For example, during the polarisation that precedes advanced stage neuroblastoma, FOXO3 – the most fibroblast migration, the nucleus moves away from the prevalent FOXO in neuronal cells – is inactivated. future leading edge of the cell. This nuclear movement involves nesprin-2G (a Notably, however, chemotherapeutic agents can activate FOXO3 and nuclear envelope spectrin repeat protein) and the SUN-domain protein SUN2, induce apoptosis in neuronal tumour cells. Judith Hagenbuchner and which are components of the linker of nucleoskeleton and cytoskeleton (LINC) colleagues (p. 1191), who have been investigating the molecular events that complex that form transmembrane actin-associated nuclear (TAN) lines at the underlie FOXO3-induced apoptosis, now report that, in two neuroblastoma nuclear envelope. Now, on page 1099, Edgar Gomes and colleagues report that cell lines, FOXO3 represses mitochondrial respiration and induces cellular the nuclear envelope protein spindle-associated membrane protein 1 (Samp1, ROS in response to chemotherapy. They show that etoposide- and also known as NET5 and TMEM201) is also required for nuclear movement in doxorubicin-induced elevation of cellular ROS depends on FOXO3 migrating fibroblasts. By using siRNA knockdown, the authors show that activation and on induction of its transcriptional target Bim (BCL2L11), a Samp1 is involved in centrosome orientation and nuclear movement during pro-apoptotic protein. -

Caveolae and Cancer: a New Mechanical Perspective
Special Edition 367 Caveolae and Cancer: A New Mechanical Perspective Christophe Lamaze1,2,3, Stéphanie Torrino1,2,3 Caveolae are small invaginations of the plasma membrane in cells. In addition to their classically described functions in cell signaling and membrane trafficking, it was recently shown that caveolae act also as plasma membrane sensors that respond immediately to acute mechanical stresses. Caveolin 1 (Cav1), the main component of caveolae, is a multifunctional scaffolding protein that can remodel the extracellular environment. Caveolae dysfunction, due to mutations in caveolins, has been linked to several human diseases called “caveolinopathies,” including muscular dystrophies, cardiac disease, infection, osteoporosis, and cancer. The role of caveolae and/or Cav1 remains controversial particularly in tumor progression. Cav1 function has been associated with several steps of cancerogenesis such as tumor growth, cell migration, metastasis, and angiogenesis, yet it was observed that Cav1 could affect these steps in a Dr. Christophe Lamaze positive or negative manner. Here, we discuss the possible function of caveolae and Cav1 in tumor progression in the context of their recently discovered role in cell mechanics. (Biomed J 2015;38:367-379) Key words: cancer, caveolae, caveolin, cavin, mechanics aveolae (for “little caves”) are small (50–100 nm) caveolins may control intracellular signaling.[8] Cplasma membrane invaginations discovered by electron The modulation of cell signaling by caveolae and/or Cav1 microscopy in 1953.[1] Caveolin (Cav) and Cavin proteins are could be one of the mechanisms by which caveolae play a the key components of caveolae that are enriched in glyco‑ role in cell transformation and tumor progression. -
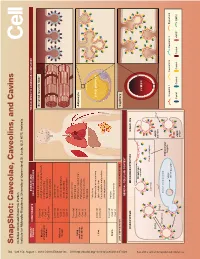
Snapshot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G
704 Cell SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton 154 Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia , August 1,2013©2013 Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.07.009 ORGAN/ COMPONENTS DISEASE AND TISSUE-SPECIFIC CAVEOLAR COMPLEXES TISSUE CELLULAR PROCESS Rippling muscle disease Caveolin3 Striated muscle fiber Limb girdle muscular dystrophy Muscle Cavin1 skeletal and Cardiomyopathy Cavin4 cardiac Mechanoprotection Cav1 (heart) T-tubule formation Caveolin1 Lipodystrophy Caveolin2 Lipotoxicity Adipose Cavin1 Fatty acid regulation tissue Cavin2 Insulin signaling Cavin3 Mechanoprotection Atherosclerosis Caveolin1 Inammation Caveolin2 Lung Pulmonary hypertension and other Cavin1 Adipocyte Pulmonary brosis endothelia Cavin2 Mechanosensation Cavin3 Signaling Fatty liver Hepatocellular carcinoma Caveolin1 LIPID DROPLET Liver Lipid metabolism Caveolin2 Carbohydrate metabolism Liver regeneration Caveolin1 Autism Brain Cavin1 Schizophrenia Cavin3 LOW EXPRESSION HIGH EXPRESSION Capillary GENERAL CELLULAR CONTEXT ENDOCYTOSIS MECHANOPROTECTION SIGNALING See online version for legend and references. and legend for version online See Membrane stretch LUMEN Cavin complex Caveola EHD2 eNOS Ca2+ inactive Rab5 Dynamin2 EARLY ENDOSOME Intracellular targets MVB/ Caveolin 1 Caveolin 2 Caveolin 3 Dynamin2 late endosome eNOS Cavin1 Cavin2 Cavin3 Cavin4 eNOS EHD2 active SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia Caveolae, submicroscopic bulb-shaped plasma membrane pits, are an abundant feature of many mammalian cells (Parton and del Pozo, 2013). Caveolae and the major proteins of caveolae, caveolins (Rothberg et al., 1992), and cavins (Hill et al., 2008), are linked to a number of human diseases such as muscular dystrophy, cardiomyopathy, and lipodys- trophy (see Glossary) (Bruno et al., 1993; Fernández et al., 2006; Hayashi et al., 2009).