Comprehensive Analysis of the Tumor Microenvironment in Cutaneous Melanoma Associated with Immune Infiltration Pan Wang1,*, Xiny
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -

1 Supplemental Material Maresin 1 Activates LGR6 Receptor
Supplemental Material Maresin 1 Activates LGR6 Receptor Promoting Phagocyte Immunoresolvent Functions Nan Chiang, Stephania Libreros, Paul C. Norris, Xavier de la Rosa, Charles N. Serhan Center for Experimental Therapeutics and Reperfusion Injury, Department of Anesthesiology, Perioperative and Pain Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts 02115, USA. 1 Supplemental Table 1. Screening of orphan GPCRs with MaR1 Vehicle Vehicle MaR1 MaR1 mean RLU > GPCR ID SD % Activity Mean RLU Mean RLU + 2 SD Mean RLU Vehicle mean RLU+2 SD? ADMR 930920 33283 997486.5381 863760 -7% BAI1 172580 18362 209304.1828 176160 2% BAI2 26390 1354 29097.71737 26240 -1% BAI3 18040 758 19555.07976 18460 2% CCRL2 15090 402 15893.6583 13840 -8% CMKLR2 30080 1744 33568.954 28240 -6% DARC 119110 4817 128743.8016 126260 6% EBI2 101200 6004 113207.8197 105640 4% GHSR1B 3940 203 4345.298244 3700 -6% GPR101 41740 1593 44926.97349 41580 0% GPR103 21413 1484 24381.25067 23920 12% NO GPR107 366800 11007 388814.4922 360020 -2% GPR12 77980 1563 81105.4653 76260 -2% GPR123 1485190 46446 1578081.986 1342640 -10% GPR132 860940 17473 895885.901 826560 -4% GPR135 18720 1656 22032.6827 17540 -6% GPR137 40973 2285 45544.0809 39140 -4% GPR139 438280 16736 471751.0542 413120 -6% GPR141 30180 2080 34339.2307 29020 -4% GPR142 105250 12089 129427.069 101020 -4% GPR143 89390 5260 99910.40557 89380 0% GPR146 16860 551 17961.75617 16240 -4% GPR148 6160 484 7128.848113 7520 22% YES GPR149 50140 934 52008.76073 49720 -1% GPR15 10110 1086 12282.67884 -

Genetic and Clinical Characteristics of Japanese Patients with Sporadic Somatotropinoma
2016, 63 (11), 953-963 Original Genetic and clinical characteristics of Japanese patients with sporadic somatotropinoma Ryusaku Matsumoto1), Masako Izawa2), Hidenori Fukuoka3), Genzo Iguchi3), Yukiko Odake1), Kenichi Yoshida1), Hironori Bando1), Kentaro Suda1), Hitoshi Nishizawa1), Michiko Takahashi1), Naoko Inoshita4), Shozo Yamada5), Wataru Ogawa1) and Yutaka Takahashi1) 1) Division of Diabetes and Endocrinology, Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan 2) Department of Pediatric Endocrinology and Metabolism, Aichi Children’s Health and Medical Center, Obu, Japan 3) Division of Diabetes and Endocrinology, Kobe University Hospital, Kobe, Japan 4) Department of Pathology, Toranomon Hospital, Tokyo, Japan 5) Department of Hypothalamic and Pituitary Surgery, Toranomon Hospital, Tokyo, Japan Abstract. Most of acromegaly is caused by a sporadic somatotropinoma and a couple of novel gene mutations responsible for somatotropinoma have recently been reported. To determine the cause of sporadic somatotropinoma in Japanese patients, we analyzed 61 consecutive Japanese patients with somatotropinoma without apparent family history. Comprehensive genetic analysis revealed that 31 patients harbored guanine nucleotide-binding protein, alpha stimulating (GNAS) mutations (50.8%) and three patients harbored aryl hydrocarbon receptor interacting protein (AIP) mutations (4.9%). No patients had G protein-coupled receptor 101 (GPR101) mutations. The patients in this cohort study were categorized into three groups of AIP, GNAS, and others and compared the clinical characteristics. The AIP group exhibited significantly younger age at diagnosis, larger tumor, and higher nadir GH during oral glucose tolerance test. In all patients with AIP mutation, macro- and invasive tumor was detected and repetitive surgery or postoperative medical therapy was needed. One case showed a refractory response to postoperative somatostatin analogue (SSA) but after the addition of cabergoline as combined therapy, serum IGF-I levels were controlled. -

1 1 2 3 Cell Type-Specific Transcriptomics of Hypothalamic
1 2 3 4 Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to 5 weight-loss 6 7 Fredrick E. Henry1,†, Ken Sugino1,†, Adam Tozer2, Tiago Branco2, Scott M. Sternson1,* 8 9 1Janelia Research Campus, Howard Hughes Medical Institute, 19700 Helix Drive, Ashburn, VA 10 20147, USA. 11 2Division of Neurobiology, Medical Research Council Laboratory of Molecular Biology, 12 Cambridge CB2 0QH, UK 13 14 †Co-first author 15 *Correspondence to: [email protected] 16 Phone: 571-209-4103 17 18 Authors have no competing interests 19 1 20 Abstract 21 Molecular and cellular processes in neurons are critical for sensing and responding to energy 22 deficit states, such as during weight-loss. AGRP neurons are a key hypothalamic population 23 that is activated during energy deficit and increases appetite and weight-gain. Cell type-specific 24 transcriptomics can be used to identify pathways that counteract weight-loss, and here we 25 report high-quality gene expression profiles of AGRP neurons from well-fed and food-deprived 26 young adult mice. For comparison, we also analyzed POMC neurons, an intermingled 27 population that suppresses appetite and body weight. We find that AGRP neurons are 28 considerably more sensitive to energy deficit than POMC neurons. Furthermore, we identify cell 29 type-specific pathways involving endoplasmic reticulum-stress, circadian signaling, ion 30 channels, neuropeptides, and receptors. Combined with methods to validate and manipulate 31 these pathways, this resource greatly expands molecular insight into neuronal regulation of 32 body weight, and may be useful for devising therapeutic strategies for obesity and eating 33 disorders. -

MRGPRX4 Is a Novel Bile Acid Receptor in Cholestatic Itch Huasheng Yu1,2,3, Tianjun Zhao1,2,3, Simin Liu1, Qinxue Wu4, Omar
bioRxiv preprint doi: https://doi.org/10.1101/633446; this version posted May 9, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 MRGPRX4 is a novel bile acid receptor in cholestatic itch 2 Huasheng Yu1,2,3, Tianjun Zhao1,2,3, Simin Liu1, Qinxue Wu4, Omar Johnson4, Zhaofa 3 Wu1,2, Zihao Zhuang1, Yaocheng Shi5, Renxi He1,2, Yong Yang6, Jianjun Sun7, 4 Xiaoqun Wang8, Haifeng Xu9, Zheng Zeng10, Xiaoguang Lei3,5, Wenqin Luo4*, Yulong 5 Li1,2,3* 6 7 1State Key Laboratory of Membrane Biology, Peking University School of Life 8 Sciences, Beijing 100871, China 9 2PKU-IDG/McGovern Institute for Brain Research, Beijing 100871, China 10 3Peking-Tsinghua Center for Life Sciences, Beijing 100871, China 11 4Department of Neuroscience, Perelman School of Medicine, University of 12 Pennsylvania, Philadelphia, PA 19104, USA 13 5Department of Chemical Biology, College of Chemistry and Molecular Engineering, 14 Peking University, Beijing 100871, China 15 6Department of Dermatology, Peking University First Hospital, Beijing Key Laboratory 16 of Molecular Diagnosis on Dermatoses, Beijing 100034, China 17 7Department of Neurosurgery, Peking University Third Hospital, Peking University, 18 Beijing, 100191, China 19 8State Key Laboratory of Brain and Cognitive Science, CAS Center for Excellence in 20 Brain Science and Intelligence Technology (Shanghai), Institute of Biophysics, 21 Chinese Academy of Sciences, Beijing, 100101, China 22 9Department of Liver Surgery, Peking Union Medical College Hospital, Chinese bioRxiv preprint doi: https://doi.org/10.1101/633446; this version posted May 9, 2019. -

GPR101 Drives Growth Hormone Hypersecretion and Gigantism in Mice Via Constitutive Activation of Gs and Gq/11
ARTICLE https://doi.org/10.1038/s41467-020-18500-x OPEN GPR101 drives growth hormone hypersecretion and gigantism in mice via constitutive activation of Gs and Gq/11 Dayana Abboud 1, Adrian F. Daly 2, Nadine Dupuis1, Mohamed Ali Bahri3, Asuka Inoue4, Andy Chevigné 5, ✉ ✉ Fabien Ectors6, Alain Plenevaux3, Bernard Pirotte 7, Albert Beckers 2 & Julien Hanson 1,7 1234567890():,; Growth hormone (GH) is a key modulator of growth and GH over-secretion can lead to gigantism. One form is X-linked acrogigantism (X-LAG), in which infants develop GH- secreting pituitary tumors over-expressing the orphan G-protein coupled receptor, GPR101. The role of GPR101 in GH secretion remains obscure. We studied GPR101 signaling pathways and their effects in HEK293 and rat pituitary GH3 cell lines, human tumors and in transgenic mice with elevated somatotrope Gpr101 expression driven by the rat Ghrhr promoter (GhrhrGpr101). Here, we report that Gpr101 causes elevated GH/prolactin secretion in trans- genic GhrhrGpr101 mice but without hyperplasia/tumorigenesis. We show that GPR101 con- stitutively activates not only Gs, but also Gq/11 and G12/13, which leads to GH secretion but not proliferation. These signatures of GPR101 signaling, notably PKC activation, are also present in human pituitary tumors with high GPR101 expression. These results underline a role for GPR101 in the regulation of somatotrope axis function. 1 Laboratory of Molecular Pharmacology, GIGA-Molecular Biology of Diseases, University of Liège, Liège, Belgium. 2 Department of Endocrinology, Centre Hospitalier Universitaire de Liège, University of Liège, Liège, Belgium. 3 GIGA-CRC in vivo Imaging, University of Liège, Liège, Belgium. -
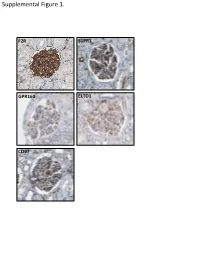
Supplemental Data
Supplemental Figure 1. F2R S1PR1 GPR160 ELTD1 CD97 Supplemental Figure 2. A B brain heart lung liver kidney spleen testis skm Expression in mouse ssues glom rok Gprc5a Gapdh Human Protein Atlas RNAseq database Supplemental Figure 3. A Gprc5a Pdgfrb Merged CL CL Gprc5a CD31 Merged CL CL B C 1.4 * 1.2 mc 1 matrix 0.8 0.6 0.4 matrix 0.2 0 pod end 1 2 mes3 D Vector Gprc5a E 40 kD - - Gprc5a - acn Supplemental Figure 4. Control 12 month-old KO 12 month-old A B C D E F 1 2 0.8 * 1.5 score 0.6 m µ 1 0.4 0.2 Slits/ 0.5 Mesangial 0 0 Ctrl KO Ctrl KO Supplemental Figure 5. A B 1.2 Vector Gprc5a 1 * 0.8 0.6 * 0.4 * Normalized Density 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 C 1.8 siCON 1.6 siGprc5a * 1.4 * * 1.2 1 0.8 0.6 NormalizedDensity 0.4 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 Supplemental table 1. List of glomerulus-expressed GPCRs as detected by qPCR. Data shown as mean ± standard deviation (Glom=glomerulus, Rok=rest of kidney). GPCR Glom Rok Glom/Rok LPAR6 41892,11 ± 38478,89 1040,06 ± 1370,12 39,28 ELTD1 30275,64 ± 14085,26 33,23 ± 46,99 910,13 GPR116 24020,06 ± 7789,84 55,1 ± 47,75 434,96 PTH1R 15402,81 ± 17644,32 7521,01 ± 3264,57 1,05 CALCRL 14096,09 ± 3854,84 199,06 ± 222,52 69,81 HPRT1 13342,04 ± 10677,69 1824,77 ± 1767,23 6,31 S1PR5 9474,7 ± 10124,3 110,84 ± 29,54 84,48 LPHN2 8645,89 ± 914,74 256,04 ± 293,87 32,77 FZD1 8176,2 ± 4947,45 1321,97 ± 1311,91 5,18 CXCR4 7097,31 ± 4388,91 535,98 ± 640,28 12,24 GPR160 6446,59 ± 1550,07 4816,3 ± 5918,99 0,34 NPY1R 6177,74 ± 6282,76 209,64 ± 296,48 28,47 PTGER4 5323,66 ± 3789,51 179,13 ± 210 28,72 RXFP1 -

Mutations in G Protein–Coupled Receptors: Mechanisms, Pathophysiology and Potential Therapeutic Approachess
Supplemental Material can be found at: /content/suppl/2020/11/26/73.1.89.DC1.html 1521-0081/73/1/89–119$35.00 https://doi.org/10.1124/pharmrev.120.000011 PHARMACOLOGICAL REVIEWS Pharmacol Rev 73:89–119, January 2021 Copyright © 2020 by The Author(s) This is an open access article distributed under the CC BY-NC Attribution 4.0 International license. ASSOCIATE EDITOR: PAUL INSEL Mutations in G Protein–Coupled Receptors: Mechanisms, Pathophysiology and Potential Therapeutic Approachess Torsten Schöneberg and Ines Liebscher Rudolf Schönheimer Institute of Biochemistry, Molecular Biochemistry, Medical Faculty, Leipzig, Germany Abstract ................................................................................... 90 Significance Statement. .................................................................. 90 I. Introduction . .............................................................................. 90 II. History .................................................................................... 92 III. General Mechanisms of GPCR Pathologies . ................................................ 93 IV. Inactivating Mutations of GPCRs .......................................................... 95 A. Partially Inactivating Mutations—Loss of Basal Activity . ............................... 97 — B. Partially Inactivating Mutations Alteration of Distinct Receptor Functions............. 97 Downloaded from C. The Special Case—Pseudogenization of GPCRs ......................................... 99 V. Activating Mutations in GPCRs—GoF..................................................... -

Update on GPCR-Based Targets for the Development of Novel Antidepressants
Molecular Psychiatry https://doi.org/10.1038/s41380-021-01040-1 EXPERT REVIEW Update on GPCR-based targets for the development of novel antidepressants 1 1 2 1 Ioannis Mantas ● Marcus Saarinen ● Zhi-Qing David Xu ● Per Svenningsson Received: 5 June 2020 / Revised: 22 January 2021 / Accepted: 25 January 2021 © The Author(s) 2021. This article is published with open access Abstract Traditional antidepressants largely interfere with monoaminergic transport or degradation systems, taking several weeks to have their therapeutic actions. Moreover, a large proportion of depressed patients are resistant to these therapies. Several atypical antidepressants have been developed which interact with G protein coupled receptors (GPCRs) instead, as direct targeting of receptors may achieve more efficacious and faster antidepressant actions. The focus of this review is to provide an update on how distinct GPCRs mediate antidepressant actions and discuss recent insights into how GPCRs regulate the pathophysiology of Major Depressive Disorder (MDD). We also discuss the therapeutic potential of novel GPCR targets, which are appealing due to their ligand selectivity, expression pattern, or pharmacological profiles. Finally, we highlight 1234567890();,: 1234567890();,: recent advances in understanding GPCR pharmacology and structure, and how they may provide new avenues for drug development. Major depressive disorder (MDD) or Major Affective Dis- because of the varied neurobiological mechanisms whereby order is a severe psychiatric disorder affecting over 200 people can qualify for a diagnosis. Moreover, patients million people worldwide [1] The life-time depression risk is who meet criteria for one mental disorder often tend to meet 15–18% and is higher in women than men [1]. -

GPR101, an Orphan GPCR with Roles in Growth and Pituitary Tumorigenesis
27 8 Endocrine-Related G Trivellin et al. Function of GPR101 27:8 T87–T97 Cancer THEMATIC REVIEW HEREDITARY ENDOCRINE TUMOURS: CURRENT STATE-OF-THE-ART AND RESEARCH OPPORTUNITIES GPR101, an orphan GPCR with roles in growth and pituitary tumorigenesis Giampaolo Trivellin1, Fabio R Faucz1, Adrian F Daly2, Albert Beckers2 and Constantine A Stratakis1 1Section on Genetics & Endocrinology (SEGEN), Intramural Research Program (IRP), Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD), Bethesda, Maryland, USA 2Department of Endocrinology, Centre Hospitalier Universitaire de Liège, University of Liège, Liège, Belgium Correspondence should be addressed to C A Stratakis: [email protected] This paper is part of a thematic section on current knowledge and future research opportunities in hereditary endocrine tumours, as discussed at MEN2019: 16th International Workshop on Multiple Endocrine Neoplasia, 27–29 March 2019, Houston, TX, USA. This meeting was sponsored by Endocrine-Related Cancer A Beckers, A F Daly and C A Stratakis are members of the editorial board of Endocrine-Related Cancer. They were not involved in the review or editorial process for this paper, on which they are listed as authors Abstract We recently described X-linked acrogigantism (X-LAG) in sporadic cases of infantile Key Words gigantism and a few familial cases of pituitary gigantism in the context of the disorder f hypothalamus known as familial isolated pituitary adenomas. X-LAG cases with early onset gigantism f pituitary (in infants or toddlers) shared copy number gains (CNG) of the distal long arm of f gigantism chromosome X (Xq26.3). In all patients described to date with Xq26.3 CNG and acro- f cyclic AMP gigantism, the only coding gene sequence shared by all chromosomal defects was that of f protein kinase A GPR101.