Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
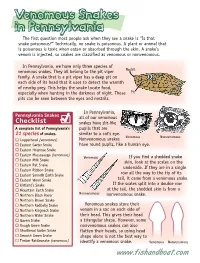
Venomous Snakes in Pennsylvania the First Question Most People Ask When They See a Snake Is “Is That Snake Poisonous?” Technically, No Snake Is Poisonous
Venomous Snakes in Pennsylvania The first question most people ask when they see a snake is “Is that snake poisonous?” Technically, no snake is poisonous. A plant or animal that is poisonous is toxic when eaten or absorbed through the skin. A snake’s venom is injected, so snakes are classified as venomous or nonvenomous. In Pennsylvania, we have only three species of venomous snakes. They all belong to the pit viper Nostril family. A snake that is a pit viper has a deep pit on each side of its head that it uses to detect the warmth of nearby prey. This helps the snake locate food, especially when hunting in the darkness of night. These Pit pits can be seen between the eyes and nostrils. In Pennsylvania, Pennsylvania Snakes all of our venomous Checklist snakes have slit-like A complete list of Pennsylvania’s pupils that are 21 species of snakes. similar to a cat’s eye. Venomous Nonvenomous Copperhead (venomous) Nonvenomous snakes Eastern Garter Snake have round pupils, like a human eye. Eastern Hognose Snake Eastern Massasauga (venomous) Venomous If you find a shedded snake Eastern Milk Snake skin, look at the scales on the Eastern Rat Snake underside. If they are in a single Eastern Ribbon Snake Eastern Smooth Earth Snake row all the way to the tip of its Eastern Worm Snake tail, it came from a venomous snake. Kirtland’s Snake If the scales split into a double row Mountain Earth Snake at the tail, the shedded skin is from a Northern Black Racer Nonvenomous nonvenomous snake. -

The Herpetology of Erie County, Pennsylvania: a Bibliography
The Herpetology of Erie County, Pennsylvania: A Bibliography Revised 2 nd Edition Brian S. Gray and Mark Lethaby Special Publication of the Natural History Museum at the Tom Ridge Environmental Center, Number 1 2 Special Publication of the Natural History Museum at the Tom Ridge Environmental Center The Herpetology of Erie County, Pennsylvania: A Bibliography Revised 2 nd Edition Compiled by Brian S. Gray [email protected] and Mark Lethaby Natural History Museum at the Tom Ridge Environmental Center, 301 Peninsula Dr., Suite 3, Erie, PA 16505 [email protected] Number 1 Erie, Pennsylvania 2017 Cover image: Smooth Greensnake, Opheodrys vernalis from Erie County, Pennsylvania. 3 Introduction Since the first edition of The herpetology of Erie County, Pennsylvania: a bibliography (Gray and Lethaby 2012), numerous articles and books have been published that are pertinent to the literature of the region’s amphibians and reptiles. The purpose of this revision is to provide a comprehensive and updated list of publications for use by researchers interested in Erie County’s herpetofauna. We have made every effort to include all major works on the herpetology of Erie County. Included are the works of Atkinson (1901) and Surface (1906; 1908; 1913) which are among the earliest to note amphibians and or reptiles specifically from sites in Erie County, Pennsylvania. The earliest publication to utilize an Erie County specimen, however, may have been that of LeSueur (1817) in his description of Graptemys geographica (Lindeman 2009). While the bibliography is quite extensive, we did not attempt to list everything, such as articles in local newspapers, and unpublished reports, although some of the more significant of these are included. -

Master Wildlife Inventory List
Wildlife Inventory List The wildlife inventory list was created from existing data, on site surveys, and/or the availability of suitable habitat. The following species could occur in the Project Area at some time during the year: Common Name Scientific Name Bird Species Bitterns, Herons, & Allies Ardeidae D, E American Bittern Botaurus lentiginosus A, E, F Great Blue Heron Ardea herodias E, F Green Heron Butorides virescens D Least bittern Ixobrychus exilis Blackbirds Icteridae B, E, F Baltimore Oriole Icterus galbula B, E, F Bobolink Dolichonyx oryzivorus B, E, F Brown-headed Cowbird Molothrus ater B, E, F Common Grackle Quiscalus quiscula B, E, F Eastern Meadowlark Sturnella magna B, E, F Red-winged Blackbird Agelaius phoeniceus Caracaras & Falcons Falconidae B, E, F, G American Kestrel Falco sparverius B Merlin Falco columbarius D Peregrine Falcon Falco peregrinus Chickadees & Titmice Paridae B, E, F, G Black-capped Chickadee Poecile atricapillus E, F, G Tufted Titmouse Baeolophus bicolor Creepers Certhiidae B, E, F Brown Creeper Certhia americana Cuckoos, Roadrunners, & Anis Cuculidae D, E, F Black-billed Cuckoo Coccyzus erythropthalmus E, F Yellow-billed Cuckoo Coccyzus americanus Finches Fringillidae B, E, F, G American Goldfinch Carduelis tristis B, E, F Chipping Sparrow Spizella passerina G Common Redpoll Acanthis flammea B, E, F Eastern Towhee Pipilo erythrophthalmus E, F, G House Finch Carpodacus mexicanus B, E Pine Siskin Spinus pinus Page 1 Eight Point Wind Energy Center E, F, G Purple Finch Carpodacus purpureus B, E Red Crossbill -

Standard Common and Current Scientific Names for North American Amphibians, Turtles, Reptiles & Crocodilians
STANDARD COMMON AND CURRENT SCIENTIFIC NAMES FOR NORTH AMERICAN AMPHIBIANS, TURTLES, REPTILES & CROCODILIANS Sixth Edition Joseph T. Collins TraVis W. TAGGart The Center for North American Herpetology THE CEN T ER FOR NOR T H AMERI ca N HERPE T OLOGY www.cnah.org Joseph T. Collins, Director The Center for North American Herpetology 1502 Medinah Circle Lawrence, Kansas 66047 (785) 393-4757 Single copies of this publication are available gratis from The Center for North American Herpetology, 1502 Medinah Circle, Lawrence, Kansas 66047 USA; within the United States and Canada, please send a self-addressed 7x10-inch manila envelope with sufficient U.S. first class postage affixed for four ounces. Individuals outside the United States and Canada should contact CNAH via email before requesting a copy. A list of previous editions of this title is printed on the inside back cover. THE CEN T ER FOR NOR T H AMERI ca N HERPE T OLOGY BO A RD OF DIRE ct ORS Joseph T. Collins Suzanne L. Collins Kansas Biological Survey The Center for The University of Kansas North American Herpetology 2021 Constant Avenue 1502 Medinah Circle Lawrence, Kansas 66047 Lawrence, Kansas 66047 Kelly J. Irwin James L. Knight Arkansas Game & Fish South Carolina Commission State Museum 915 East Sevier Street P. O. Box 100107 Benton, Arkansas 72015 Columbia, South Carolina 29202 Walter E. Meshaka, Jr. Robert Powell Section of Zoology Department of Biology State Museum of Pennsylvania Avila University 300 North Street 11901 Wornall Road Harrisburg, Pennsylvania 17120 Kansas City, Missouri 64145 Travis W. Taggart Sternberg Museum of Natural History Fort Hays State University 3000 Sternberg Drive Hays, Kansas 67601 Front cover images of an Eastern Collared Lizard (Crotaphytus collaris) and Cajun Chorus Frog (Pseudacris fouquettei) by Suzanne L. -

Volume 55, Number 1 01/01/2020
BULLETIN of the Chicago Herpetological Society Volume 55, Number 1 January 2020 BULLETIN OF THE CHICAGO HERPETOLOGICAL SOCIETY Volume 55, Number 1 January 2020 A Letter from the Outgoing CHS President . Rich Crowley 1 The Herpetofauna of Presque Isle State Park, Erie, Pennsylvania: Annotated Checklist and Comprehensive Bibliography . Brian S. Gray 2 Turtle speak? An Unusual (Unique?) Display of Mass Social Interaction among Arizona Mud Turtles (Kinosternon arizonense) in Pima County, Arizona. Roger A. Repp 19 Minutes of the CHS Board Meeting, December 13, 2019 . 22 Chicago Herpetological Society Income Statement: January 1 --- December 31, 2019, and Balance Sheet, December 31, 2019 . 23 Advertisements . 24 New CHS Members This Month . 24 Cover: Gray treefrog, Hyla versicolor, Presque Isle State Park, Erie County, Pennsylvania. Photograph by Mark Lethaby. STAFF Membership in the CHS includes a subscription to the monthly Bulletin. Annual dues are: Individual Membership, $25.00; Editor: Michael A. Dloogatch --- [email protected] Family Membership, $28.00; Sustaining Membership, $50.00; Copy editor: Joan Moore Contributing Membership, $100.00; Institutional Membership, $38.00. Remittance must be made in U.S. funds. Subscribers 2019 CHS Board of Directors outside the U.S. must add $12.00 for postage. Send membership dues or address changes to: Chicago Herpetological Society, President: John Gutierrez Membership Secretary, 2430 N. Cannon Drive, Chicago, IL 60614. Vice-president: Jessica Wadleigh Treasurer: John Archer Manuscripts published in the Bulletin of the Chicago Herpeto- Recording Secretary: Gail Oomens logical Society are not peer reviewed. Manuscripts and letters Media Secretary: Annalisa Kolb concerning editorial business should be e-mailed to the editor, Membership Secretary: Mike Dloogatch [email protected]. -

Preliminary Morphometrics, Growth, and Natural History Observations Of
RESEARCH ARTICLE The Herpetological Bulletin 141, 2017: 1-6 Preliminary morphometrics, growth, and natural history observations of the short-headed garter snake, Thamnophis brachystoma at two urban sites in Erie County, Pennsylvania, USA BRIAN S. GRAY* & MARK LETHABY Natural History Museum at the Tom Ridge Environmental Center 301 Peninsula Drive, Erie, Pennsylvania 16505, USA *Corresponding author Email: [email protected] ABSTRACT - We used mark-recapture techniques to study the short-headed garter snake, Thamnophis brachystoma at two urban sites (Shannon Road and McClelland Park) in Erie County, Pennsylvania, USA. Mean snout-vent length (SVL) and weight was greater in females than males regardless of age class; whereas relative to total length, tail length was consistently greater in males than in females. Sex ratios did not differ significantly from 1:1 regardless of age-class. At Shannon Road, adults significantly outnumbered juveniles 3.3 to 1. While at McClelland Park, juveniles (N = 63) were nearly twice as numerous as adults (N = 31). Data regarding estimated growth are also reported. The results of the present study conform to previously published data from populations in Pennsylvania and a population near Olean, New York. INTRODUCTION he short-headed garter snake, Thamnophis brachystoma (Fig. 1) is relatively small with a recorded maximum Tlength of 578 mm (Lethaby, 2004). It is found primarily in early successional habitats in the Allegheny High Plateau of north-western Pennsylvania and south-western New York, USA (Price, 1978). Introduced populations have been reported in New York (Bothner, 1976), Ohio (Novotny, 1990; Novotny et al., 2011), and Pennsylvania (Price, 1978; McCoy, 1982). -

Wildlife Site Characterization Report South Ripley Solar Town of Ripley Chautauqua County, New York
Wildlife Site Characterization Report South Ripley Solar Town of Ripley Chautauqua County, New York Prepared for: ConnectGen LLC 1001 McKinney, Suite 700 Houston, Texas 77002 Prepared by: Environmental Design & Research, Landscape Architecture, Engineering, & Environmental Services, D.P.C. 217 Montgomery Street, Suite 1000 Syracuse, New York 13202 www.edrdpc.com February 2021 TABLE OF CONTENTS 1.0 INTRODUCTION ............................................................................................................................................... 1 2.0 PUBLICLY AVAILABLE DATA SOURCES ........................................................................................................ 2 2.1 NEW YORK’S EAF MAPPER ....................................................................................................................... 2 2.2 NEW YORK’S ENVIRONMENTAL RESOURCE MAPPER .......................................................................... 2 2.3 NEW YORK NATURAL HERITAGE PROGRAM.......................................................................................... 3 2.4 USFWS IPaC and ECOS DATABASES ....................................................................................................... 3 2.5 NEW YORK’S NATURE EXPLORER ........................................................................................................... 4 2.6 NEW YORK’S BIODIVERSITY AND WIND SITING MAPPING TOOL......................................................... 4 2.7 CORNELL LABORATORY OF ORNITHOLOGY’s eBird ............................................................................. -

2010 Box Score
PENNSYLVANIA’S WILD SPECIES — THE BOX SCORE SPECIES* OF IMMEDIATE CONCERN TOTAL SPECIES (INCLUDING SUBSPECIES & VARIETIES) PRESUMED REPRODUCING CURRENTLY IN NON-BREEDING PABS- PABS- DOWNLISTED PENNSYLVANIA EXTIRPATED EXTINCT PENNSYLVANIA MIGRANTS OR GROUP OF ORGANISMS RECOMMENDED RECOMMENDED SINCE 1990 DUE RESPONSIBILITY FROM (GLOBALLY) WINTER † † TO RECOVERY ‡ ENDANGERED THREATENED PENNSYLVANIA NATIVE NONNATIVE TOTAL RESIDENTS SPECIES PLANTS Vascular plants G 24 + G 4 + G 10 + Rev. May 2010 — B. Isaac, S. Grund 0 2 2,075 931 3,006 — ? & T. Block P 300 P 115 P 97 MOSSES ~350 0 MOSSES ~350 Bryophytes ? ? 0 P 3 0 LIVERWORTS LIVERWORTS — ? Rev. Sep. 2007 — J. Lendemer ~150 known ~150 Other plant groups, whose status in Pennsylvania is less well known, are green algae (Chlorophyta) and stoneworts or pondweeds (Charophyta). ANIMALS — CHORDATES G 1 + G 2 + Birds P 3 2 2 184 5 189 196 9 Rev. June 2010 — M. Brittingham P 14 P 4 G 1 + G 2 + Mammals 1 P 9 0 63 3 65 0 4 Rev. June 2010 — M. Gannon P 3 P 1 G 2 + Reptiles P 1 0 P 2 0 35 1 36 0 7 To be rev. Aug. 2010 — T. Maret P 2 Amphibians P 4 P 1 0 P 1 0 37 0 37 0 3 To be rev. Aug. 2010 — T. Maret G 5 + G 2 + G 2 + Fishes 6 1 165 18 183 26 13 Rev. June 2010 — R. Criswell P 23 P 7 P 14 ANIMALS — ARTHROPODS G 4 + G 1 + G 1 + Butterflies 0 0 122 2 124 20 2 Rev. July 2010 — B. -

A Guide to the Reptiles of Erie County, Pennsylvania
2 A Guide to the Reptiles of Erie County, Pennsylvania BRIAN S. GRAY Natural History Museum at the Tom Ridge Environmental Center Erie, Pennsylvania, USA. Published by Brian S. Gray Copyright © 2011 Brian S. Gray 3 Table of Contents Introduction…6 Acknowledgements…9 Species accounts…10 Snakes Harmless egg-laying snakes Eastern Racer…12 Milk Snake…14 Smooth Green Snake…16 Midland Rat Snake…18 Slender rear-fanged snakes (harmless) Ringneck Snake…20 Harmless live-bearing snakes Northern Water Snake…22 Queen Snake…24 Brown Snake…26 Redbelly Snake…28 Shorthead Garter Snake…30 Eastern Ribbon Snake…32 Common Garter Snake…34 Robust rear-fanged snakes (harmless) Eastern Hognose Snake…36 Lizards Skinks Five-lined Skink…40 4 Turtles Snapping turtles Common Snapping Turtle…44 Mud and Musk Turtles Common Musk Turtle…48 Box and Basking Turtles Spotted Turtle…50 Northern Painted Turtle…52 Blanding’s Turtle…54 Wood Turtle…56 Common Map Turtle…58 Eastern Box Turtle…60 Softshells Spiny Softshell…62 Nonnative species…64 Identifying shed snakeskins…66 Glossary…75 Maps…78 Bibliography…82 5 INTRODUCTION Erie County in northwestern Pennsylvania, a formerly glaciated region, is unique among the commonwealth’s 67counties in that it is bordered by Lake Erie on its north side. The county is divided into 21 townships (Figure 1) and contains the city of Erie, the fourth largest city in Pennsylvania. Two physiographic provinces occur in the county, the Central Lowland and the Appalachian Plateaus. The Central Lowland province consists of a narrow band 2–5 miles wide extending along the Lake Erie shoreline. -

Lasher Road Substation Project Appendix E Plant and Wildlife
Lasher Road Substation Project Appendix E Plant and Wildlife Species List PLANT SPECIES INVENTORY Observed On-Site During TRC Surveys, 2015 Family Genus species common name Aceraceae Acer rubrum red maple Aceraceae Acer saccharum sugar maple Anacardiaceae Rhus typhina staghorn sumac Anacardiaceae Toxicodendron radicans eastern poison ivy Apiaceae Daucus carota wild carrot Apiaceae Pastinaca sativa wild parsnip Apocynaceae Asclepias syriaca common milkweed Asteraceae ` Ambrosia artemisiifolia annual ragweed Asteraceae Solidago altissima tall goldenrod Asteraceae Solidago rugosa wrinkled-leaved goldenrod Asteraceae Solidago spp. goldenrods Asteraceae Symphyotrichum spp. asters Balsaminaceae Impatiens capensis spotted jewelweed Betulaceae Alnus incana speckled alder Betulaceae Betula alleghaniensis yellow birch Betulaceae Betula lenta black birch Betulaceae Betula populifolia gray birch Betulaceae Carpinus caroliniana American hornbeam Brassicaceae Alliaria petiolata garlic mustard Caprifoliaceae Lonicera spp. bush honeysuckle Cornaceae Cornus sericea red osier dogwood Cyperaceae Carex lacustris lake-bank sedge Cyperaceae Carex spp. sedges Cyperaceae Scirpus spp. bulrushes Dryopteridaceae Polystichum acrostichoides Christmas fern Elaeagnaceae Elaeagnus umbellata autumn olive Equisetaceae Equisetum arvense field horsetail Fabaceae Baptisia tinctoria wild-indigo Fabaceae Robinia pseudoacacia black locust Fabaceae Trifolium pretense red clover Fagaceae Quercus alba white oak Lasher Road Substation Page 1 Family Genus species common name Fagaceae -
The Herpetological Bulletin
THE HERPETOLOGICAL BULLETIN The Herpetological Bulletin is produced quarterly and publishes, in English, a range of articles concerned with herpetology. These include society news, full-length papers, new methodologies, natural history notes, book reviews, letters from readers and other items of general herpetological interest. Emphasis is placed on natural history, conservation, captive breeding and husbandry, veterinary and behavioural aspects. Articles reporting the results of experimental research, descriptions of new taxa, or taxonomic revisions should be submitted to The Herpetological Journal (see inside back cover for Editor’s address). Guidelines for Contributing Authors: 1. See the BHS website for a free download of the Bulletin showing Bulletin style. A template is available from the BHS website www.thebhs.org or on request from the Editor. 2. Contributions should be submitted by email or as text files on CD or DVD in Windows® format using standard word-processing software. 3. Articles should be arranged in the following general order: Title Name(s) of authors(s) Address(es) of author(s) (please indicate corresponding author) Abstract (required for all full research articles - should not exceed 10% of total word length) Text acknowledgements References Appendices Footnotes should not be included. 4. Text contributions should be plain formatted with no additional spaces or tabs. It is requested that the References section is formatted following the Bulletin house style (refer to this issue as a guide to style and format). Particular attention should be given to the format of citations within the text and to references. 5. High resolution scanned images (TIFF or JPEG files) are the preferred format for illustrations, although good quality slides, colour and monochrome prints are also acceptable. -

Kansas Herpetology
ISSN 1540-773X JOURNAL OF KANSAS HERPETOLOGY NUMBER 38 JULY 2011 Published by the Kansas Herpetological Society http://www.cnah.org/khs EDItoRIAL BOARD KHS OFFICERS (2011) Associate Editors President – DEREK A. SCHMIDT JOSEPH T. COLLINS 7741 SE 101st Street Kansas Biological Survey Overbrook, Kansas 66524 785.224.7549 TRAVIS W. TAGGART [email protected] Sternberg Museum of Natural History President-Elect – TRAVIS W. TAGGART Copy Editor Sternberg Museum of Natural History DANIEL G. MURROW 3000 Sternberg Drive Hutchinson, Kansas Hays, Kansas 67601-2006 Article Editors 785.650.2445 EVA HORNE [email protected] Kansas State University Past-President – KathY ELLIS GEORGE R. PISANI 10025 SW Jordan Road Kansas Biological Survey Wakarusa, Kansas 66546 LYNNEttE SIEVERT 785.383.2788 Emporia State University [email protected] WILLIAM StaRK Fort Hays State University Treasurer – DAVID OLDHAM JAMES TRIPLEtt 716 Michigan Street Pittsburg State University Oswego, Kansas 316.795.2293 [email protected] LIAISON REPRESEntatIVES Secretary – EVA A. HORNE Kansas Department of Wildlife and Parks Division of Biology KEN BRUNSON Kansas State University 620.672-5911 Manhattan, Kansas 66506 785.532.5929 Kansas Nongame Wildlife Advisory Council [email protected] TRAVIS W. TAGGART 785.650.2445 Historian – SUZANNE L. COLLINS The Center for North American Herpetology Kansas Chapter – Wildlife Society 1502 Medinah Circle CURTIS J. SCHMIDT Lawrence, Kansas 66047 785.650.2447 785.393.2392 [email protected] DISTINGUISHED LIFE MEMBERS Editor – CURTIS J. SCHMIDT ROBERT F. CLARKE Sternberg Museum of Natural History Emporia State University, Emporia, Kansas 3000 Sternberg Drive (1919–2008) Hays, Kansas 67601-2006 785.650.2447 JOSEPH T. COLLINS [email protected] Museum of Natural History, The University of Kansas Lawrence, Kansas HENRY S.